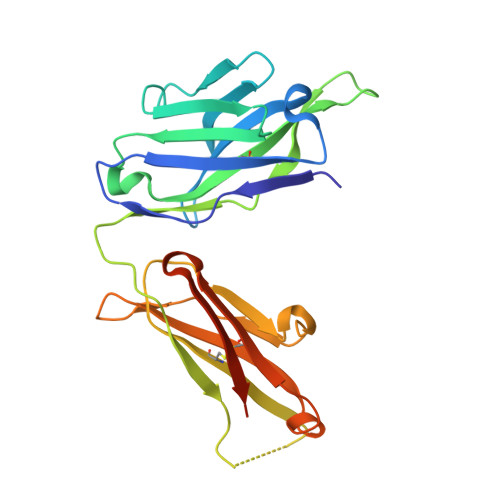
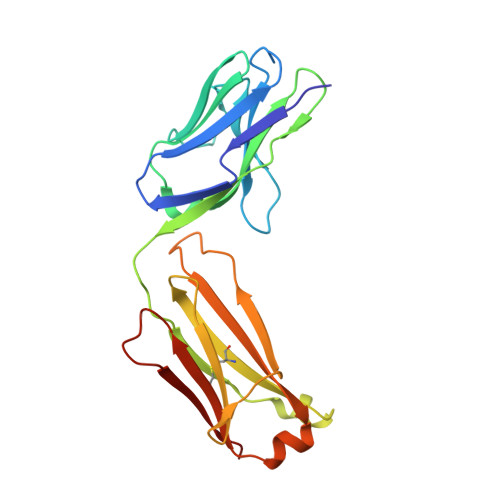
A single T cell epitope drives the neutralizing anti-drug antibody response to natalizumab in multiple sclerosis patients.
Cassotta, A., Mikol, V., Bertrand, T., Pouzieux, S., Le Parc, J., Ferrari, P., Dumas, J., Auer, M., Deisenhammer, F., Gastaldi, M., Franciotta, D., Silacci-Fregni, C., Fernandez Rodriguez, B., Giacchetto-Sasselli, I., Foglierini, M., Jarrossay, D., Geiger, R., Sallusto, F., Lanzavecchia, A., Piccoli, L.(2019) Nat Med 25: 1402-1407
- PubMed: 31501610
- DOI: https://doi.org/10.1038/s41591-019-0568-2
- Primary Citation of Related Structures:
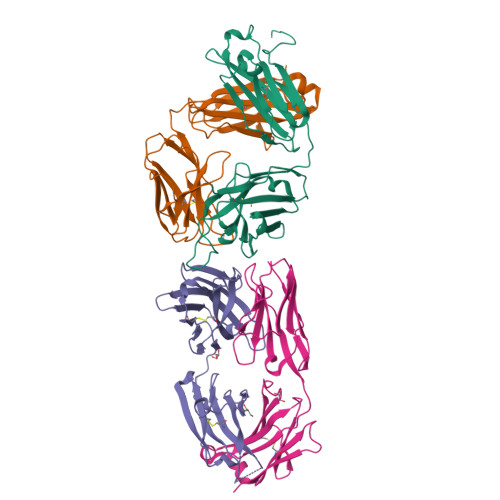
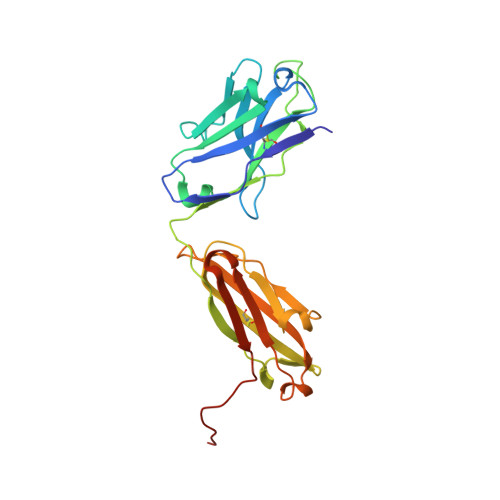
6FG1, 6FG2 - PubMed Abstract:
Natalizumab (NZM), a humanized monoclonal IgG4 antibody to α4 integrins, is used to treat patients with relapsing-remitting multiple sclerosis (MS) 1,2 , but in about 6% of the cases persistent neutralizing anti-drug antibodies (ADAs) are induced leading to therapy discontinuation 3,4 . To understand the basis of the ADA response and the mechanism of ADA-mediated neutralization, we performed an in-depth analysis of the B and T cell responses in two patients. By characterizing a large panel of NZM-specific monoclonal antibodies, we found that, in both patients, the response was polyclonal and targeted different epitopes of the NZM idiotype. The neutralizing activity was acquired through somatic mutations and correlated with a slow dissociation rate, a finding that was supported by structural data. Interestingly, in both patients, the analysis of the CD4 + T cell response, combined with mass spectrometry-based peptidomics, revealed a single immunodominant T cell epitope spanning the FR2-CDR2 region of the NZM light chain. Moreover, a CDR2-modified version of NZM was not recognized by T cells, while retaining binding to α4 integrins. Collectively, our integrated analysis identifies the basis of T-B collaboration that leads to ADA-mediated therapeutic resistance and delineates an approach to design novel deimmunized antibodies for autoimmune disease and cancer treatment.
Organizational Affiliation:
Institute for Research in Biomedicine, Università della Svizzera italiana, Bellinzona, Switzerland.