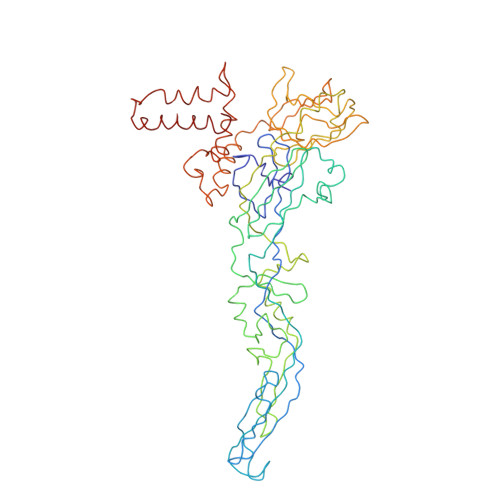
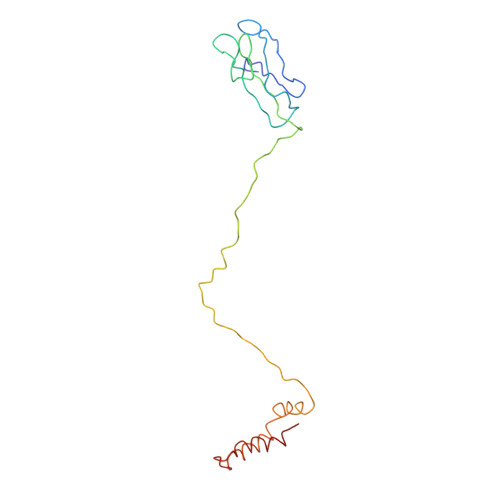
Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Tan, T.Y., Fibriansah, G., Kostyuchenko, V.A., Ng, T.S., Lim, X.X., Zhang, S., Lim, X.N., Wang, J., Shi, J., Morais, M.C., Corti, D., Lok, S.M.(2020) Nat Commun 11: 895-895
- PubMed: 32060358
- DOI: https://doi.org/10.1038/s41467-020-14647-9
- Primary Citation of Related Structures:
6LNT, 6LNU - PubMed Abstract:
Structures of flavivirus (dengue virus and Zika virus) particles are known to near-atomic resolution and show detailed structure and arrangement of their surface proteins (E and prM in immature virus or M in mature virus). By contrast, the arrangement of the capsid proteins:RNA complex, which forms the core of the particle, is poorly understood, likely due to inherent dynamics. Here, we stabilize immature Zika virus via an antibody that binds across the E and prM proteins, resulting in a subnanometer resolution structure of capsid proteins within the virus particle. Fitting of the capsid protein into densities shows the presence of a helix previously thought to be removed via proteolysis. This structure illuminates capsid protein quaternary organization, including its orientation relative to the lipid membrane and the genomic RNA, and its interactions with the transmembrane regions of the surface proteins. Results show the capsid protein plays a central role in the flavivirus assembly process.
Organizational Affiliation:
Programme in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore, 169857, Singapore.