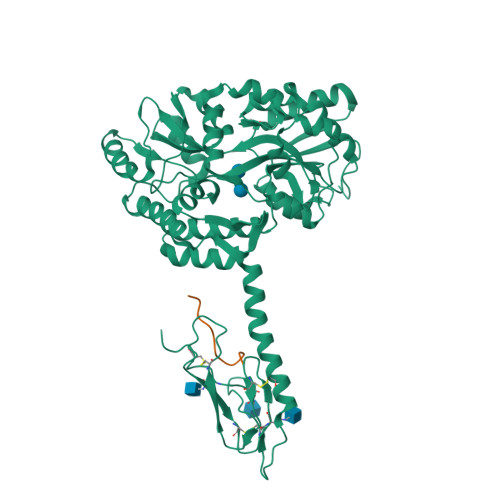
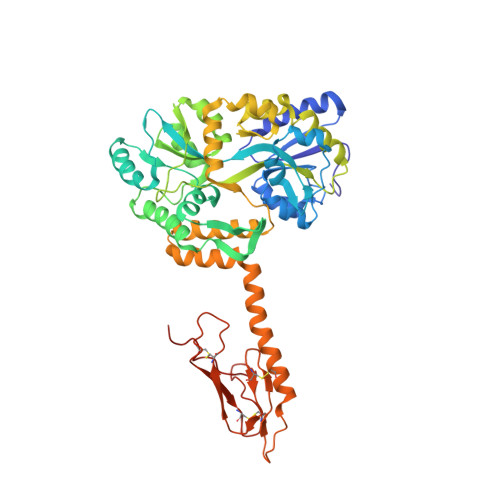
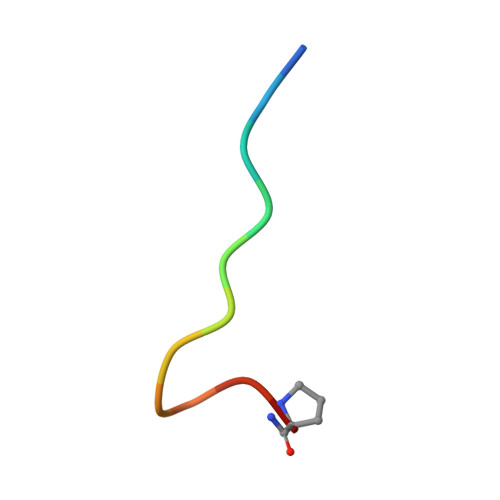
Calcitonin Receptor N-Glycosylation Enhances Peptide Hormone Affinity by Controlling Receptor Dynamics.
Lee, S.M., Jeong, Y., Simms, J., Warner, M.L., Poyner, D.R., Chung, K.Y., Pioszak, A.A.(2020) J Mol Biology 432: 1996-2014
- PubMed: 32035902
- DOI: https://doi.org/10.1016/j.jmb.2020.01.028
- Primary Citation of Related Structures:
6PFO, 6PGQ - PubMed Abstract:
The class B G protein-coupled receptor (GPCR) calcitonin receptor (CTR) is a drug target for osteoporosis and diabetes. N-glycosylation of asparagine 130 in its extracellular domain (ECD) enhances calcitonin hormone affinity with the proximal GlcNAc residue mediating this effect through an unknown mechanism. Here, we present two crystal structures of salmon calcitonin-bound, GlcNAc-bearing CTR ECD at 1.78 and 2.85 Å resolutions and analyze the mechanism of the glycan effect. The N130 GlcNAc does not contact the hormone. Surprisingly, the structures are nearly identical to a structure of hormone-bound, N-glycan-free ECD, which suggested that the GlcNAc might affect CTR dynamics not observed in the static crystallographic snapshots. Hydrogen-deuterium exchange mass spectrometry and molecular dynamics simulations revealed that glycosylation stabilized a β-sheet adjacent to the N130 GlcNAc and the N-terminal α-helix near the peptide-binding site while increasing flexibility of the peptide-binding site turret loop. These changes due to N-glycosylation increased the ligand on-rate and decreased its off-rate. The glycan effect extended to RAMP-CTR amylin receptor complexes and was also conserved in the related CGRP receptor. These results reveal that N-glycosylation can modulate GPCR function by altering receptor dynamics.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, 73104, USA; Present Address: Department of Basic Pharmaceutical Sciences, Fred Wilson School of Pharmacy, High Point University, High Point, NC, 27268, USA.