Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
Lunk, I., Litty, F.A., Hennig, S., Vetter, I.R., Kotthaus, J., Altmann, K.S., Ott, G., Havemeyer, A., Carillo Garcia, C., Clement, B., Schade, D.(2020) J Med Chem 63: 425-432
- PubMed: 31841335
- DOI: https://doi.org/10.1021/acs.jmedchem.9b01230
- Primary Citation of Related Structures:
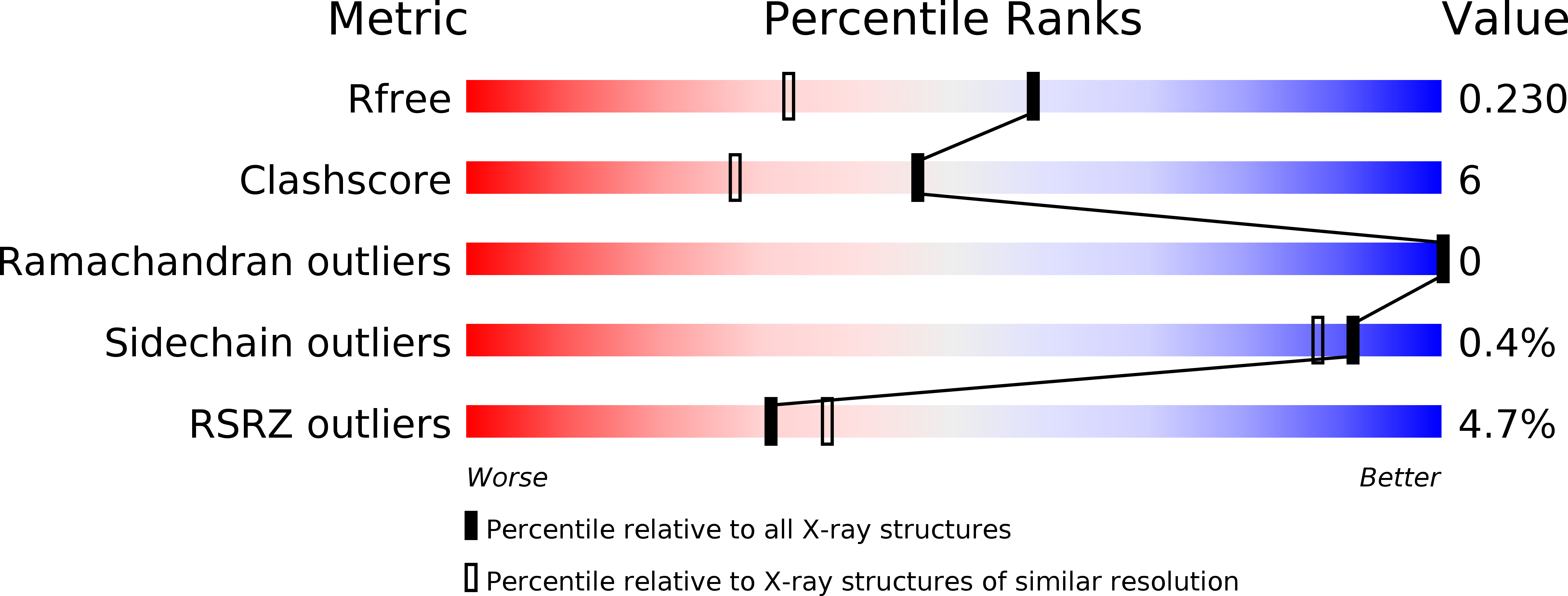
6SZP, 6SZQ - PubMed Abstract:
N -(4-Aminobutyl)- N '-(2-methoxyethyl)guanidine ( 8a ) is a potent inhibitor targeting the h DDAH-1 active site ( K i = 18 μM) and derived from a series of guanidine- and amidine-based inhibitors. Its nonamino acid nature leads to high selectivities toward other enzymes of the nitric oxide-modulating system. Crystallographic data of 8a -bound h DDAH-1 illuminated a unique binding mode. Together with its developed N -hydroxyguanidine prodrug 11 , 8a will serve as a most widely applicable, pharmacological tool to target DDAH-1-associated diseases.
Organizational Affiliation:
Department of Pharmaceutical and Medicinal Chemistry , Christian-Albrechts-University Kiel , Gutenbergstrasse 76 , D-24118 Kiel , Germany.