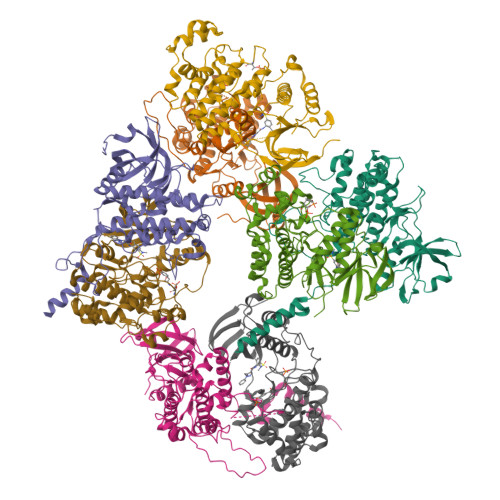
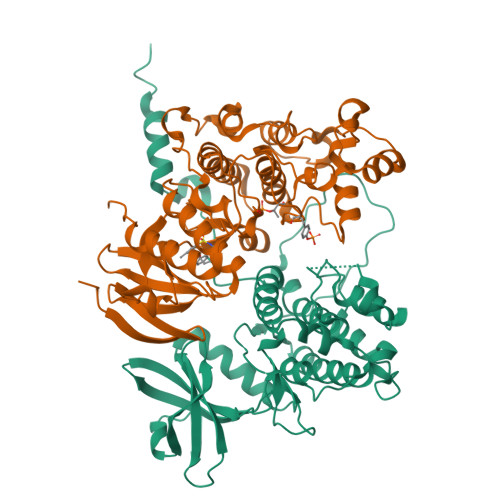
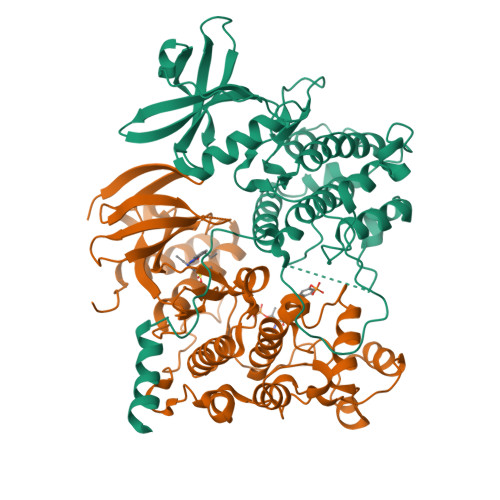
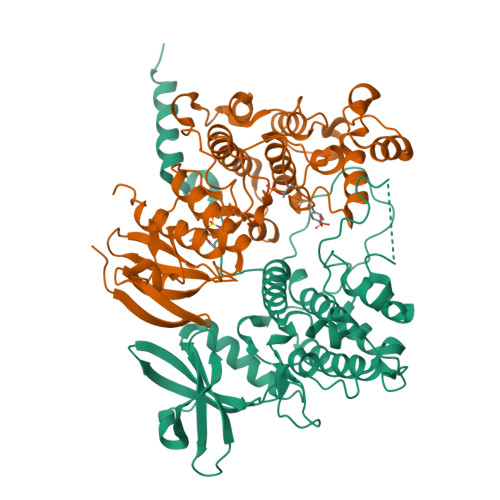
MAP Kinase-Mediated Activation of RSK1 and MK2 Substrate Kinases.
Sok, P., Gogl, G., Kumar, G.S., Alexa, A., Singh, N., Kirsch, K., Sebo, A., Drahos, L., Gaspari, Z., Peti, W., Remenyi, A.(2020) Structure 28: 1101-1113.e5
- PubMed: 32649858
- DOI: https://doi.org/10.1016/j.str.2020.06.007
- Primary Citation of Related Structures:
6TCA - PubMed Abstract:
Mitogen-activated protein kinases (MAPKs) control essential eukaryotic signaling pathways. While much has been learned about MAPK activation, much less is known about substrate recruitment and specificity. MAPK substrates may be other kinases that are crucial to promote a further diversification of the signaling outcomes. Here, we used a variety of molecular and cellular tools to investigate the recruitment of two substrate kinases, RSK1 and MK2, to three MAPKs (ERK2, p38α, and ERK5). Unexpectedly, we identified that kinase heterodimers form structurally and functionally distinct complexes depending on the activation state of the MAPK. These may be incompatible with downstream signaling, but naturally they may also form structures that are compatible with the phosphorylation of the downstream kinase at the activation loop, or alternatively at other allosteric sites. Furthermore, we show that small-molecule inhibitors may affect the quaternary arrangement of kinase heterodimers and thus influence downstream signaling in a specific manner.
Organizational Affiliation:
Biomolecular Interactions Research Group, Institute of Organic Chemistry, Research Center for Natural Sciences, Magyar Tudósok körútja 2., 1117 Budapest, Hungary.