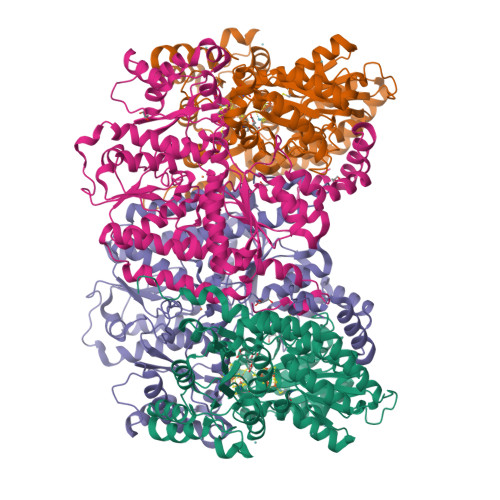
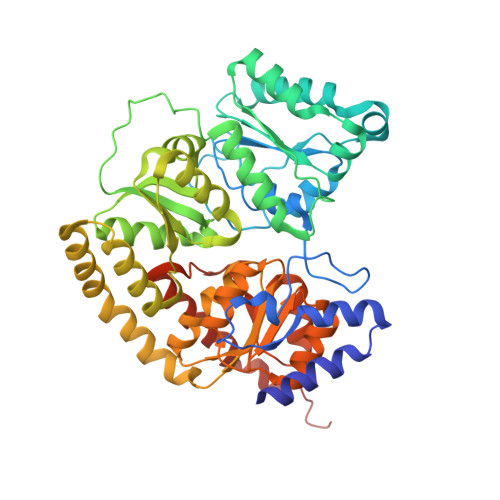
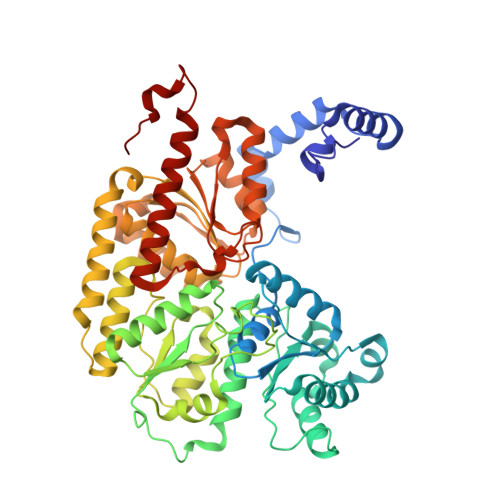
Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Kang, W., Lee, C.C., Jasniewski, A.J., Ribbe, M.W., Hu, Y.(2020) Science 368: 1381-1385
- PubMed: 32554596
- DOI: https://doi.org/10.1126/science.aaz6748
- Primary Citation of Related Structures:
6UG0, 6VXT - PubMed Abstract:
The enzyme nitrogenase uses a suite of complex metallocofactors to reduce dinitrogen (N 2 ) to ammonia. Mechanistic details of this reaction remain sparse. We report a 1.83-angstrom crystal structure of the nitrogenase molybdenum-iron (MoFe) protein captured under physiological N 2 turnover conditions. This structure reveals asymmetric displacements of the cofactor belt sulfurs (S2B or S3A and S5A) with distinct dinitrogen species in the two αβ dimers of the protein. The sulfur-displaced sites are distinct in the ability of protein ligands to donate protons to the bound dinitrogen species, as well as the elongation of either the Mo-O5 (carboxyl) or Mo-O7 (hydroxyl) distance that switches the Mo-homocitrate ligation from bidentate to monodentate. These results highlight the dynamic nature of the cofactor during catalysis and provide evidence for participation of all belt-sulfur sites in this process.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, CA 92697-3900, USA.