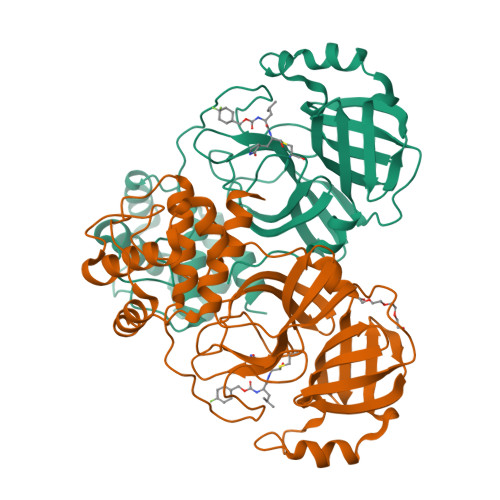
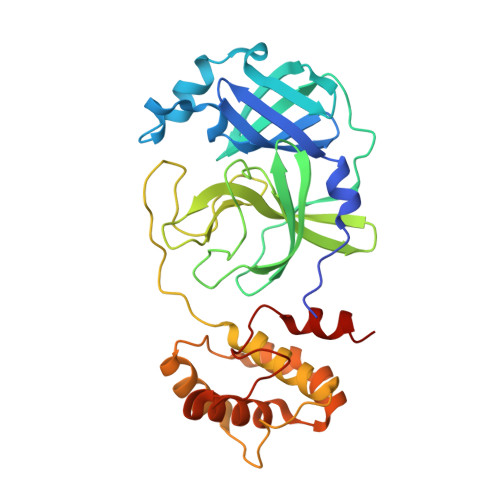
3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Rathnayake, A.D., Zheng, J., Kim, Y., Perera, K.D., Mackin, S., Meyerholz, D.K., Kashipathy, M.M., Battaile, K.P., Lovell, S., Perlman, S., Groutas, W.C., Chang, K.O.(2020) Sci Transl Med 12
- PubMed: 32747425
- DOI: https://doi.org/10.1126/scitranslmed.abc5332
- Primary Citation of Related Structures:
6VGY, 6VGZ, 6VH0, 6VH1, 6VH2, 6VH3, 6W2A, 6XMK - PubMed Abstract:
Pathogenic coronaviruses are a major threat to global public health, as exemplified by severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and the newly emerged SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19). We describe herein the structure-guided optimization of a series of inhibitors of the coronavirus 3C-like protease (3CLpro), an enzyme essential for viral replication. The optimized compounds were effective against several human coronaviruses including MERS-CoV, SARS-CoV, and SARS-CoV-2 in an enzyme assay and in cell-based assays using Huh-7 and Vero E6 cell lines. Two selected compounds showed antiviral effects against SARS-CoV-2 in cultured primary human airway epithelial cells. In a mouse model of MERS-CoV infection, administration of a lead compound 1 day after virus infection increased survival from 0 to 100% and reduced lung viral titers and lung histopathology. These results suggest that this series of compounds has the potential to be developed further as antiviral drugs against human coronaviruses.
Organizational Affiliation:
Department of Chemistry, Wichita State University, Wichita, KS 67260, USA.