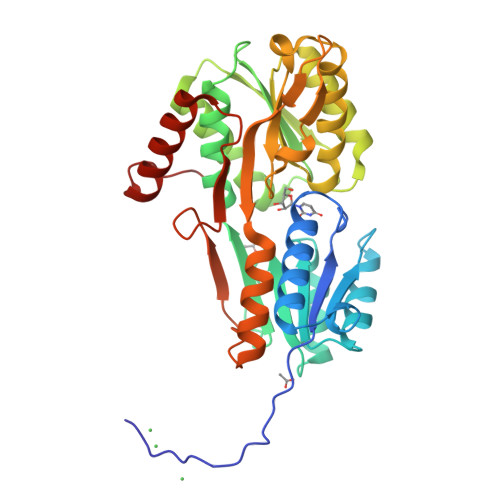
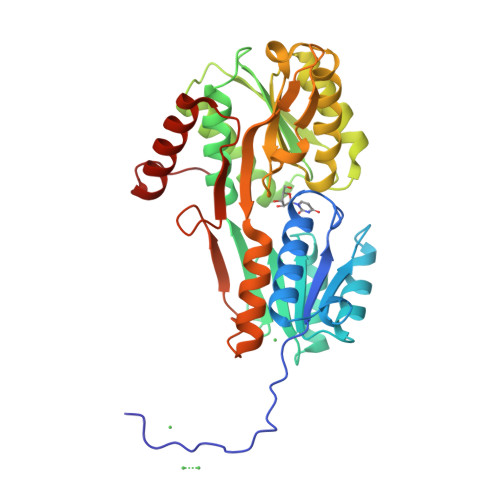
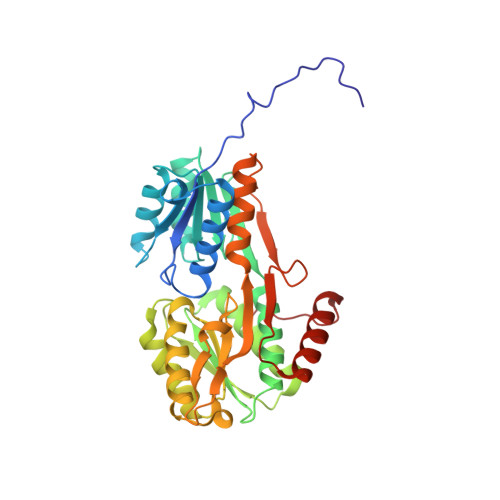
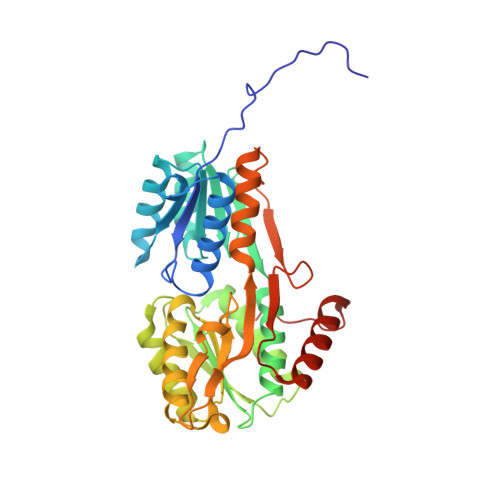
Crystal Structure and Pathophysiological Role of the Pneumococcal Nucleoside-binding Protein PnrA.
Abdullah, M.R., Batuecas, M.T., Jennert, F., Voss, F., Westhoff, P., Kohler, T.P., Molina, R., Hirschmann, S., Lalk, M., Hermoso, J.A., Hammerschmidt, S.(2021) J Mol Biology 433: 166723-166723
- PubMed: 33242497
- DOI: https://doi.org/10.1016/j.jmb.2020.11.022
- Primary Citation of Related Structures:
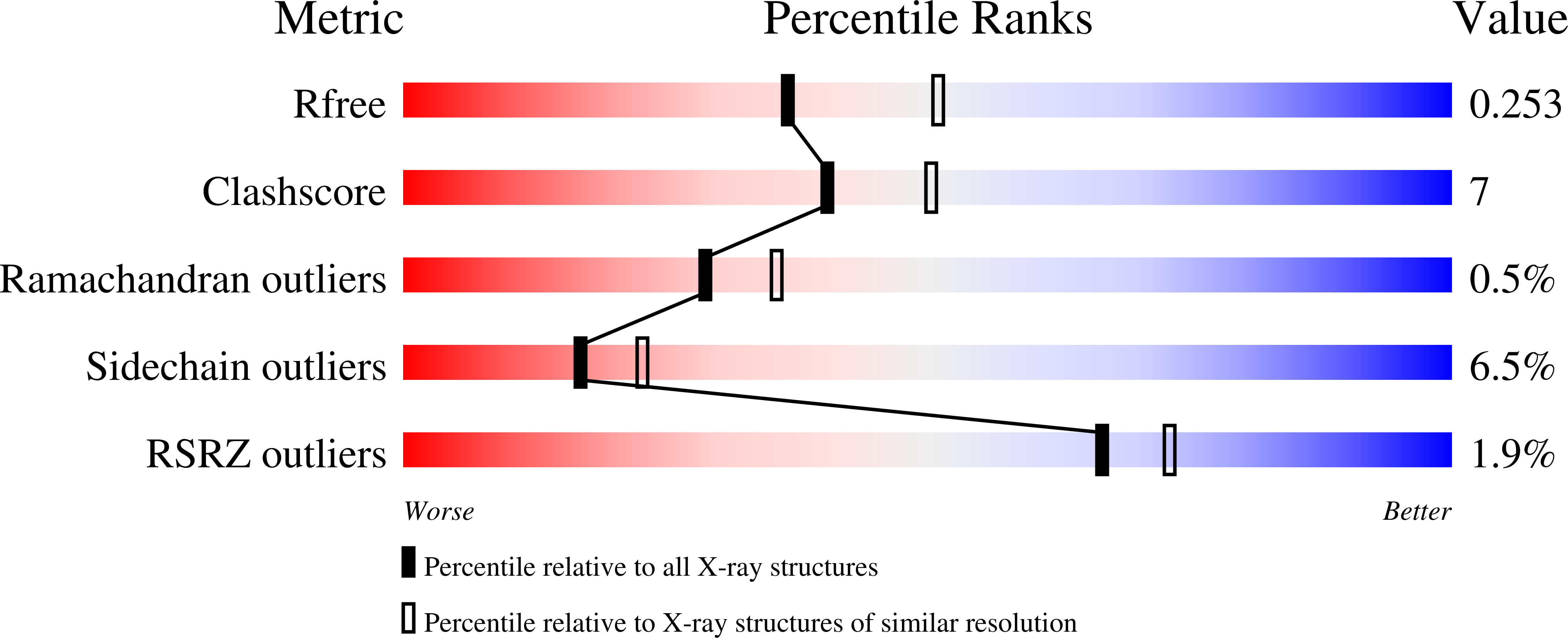
6Y9U, 6YA3, 6YA4, 6YAB, 6YAG - PubMed Abstract:
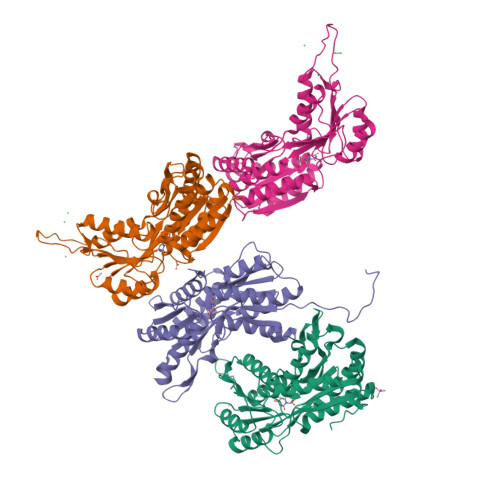
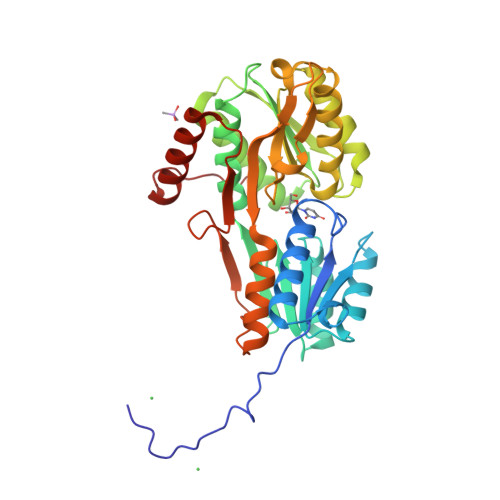
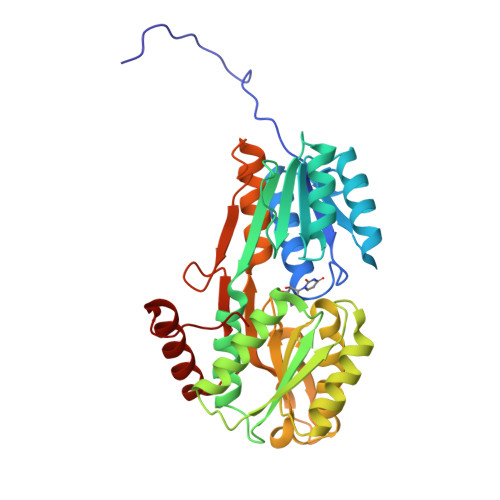
Nucleotides are important for RNA and DNA synthesis and, despite a de novo synthesis by bacteria, uptake systems are crucial. Streptococcus pneumoniae, a facultative human pathogen, produces a surface-exposed nucleoside-binding protein, PnrA, as part of an ABC transporter system. Here we demonstrate the binding affinity of PnrA to nucleosides adenosine, guanosine, cytidine, thymidine and uridine by microscale thermophoresis and indicate the consumption of adenosine and guanosine by 1 H NMR spectroscopy. In a series of five crystal structures we revealed the PnrA structure and provide insights into how PnrA can bind purine and pyrimidine ribonucleosides but with preference for purine ribonucleosides. Crystal structures of PnrA:nucleoside complexes unveil a clear pattern of interactions in which both the N- and C- domains of PnrA contribute. The ribose moiety is strongly recognized through a conserved network of H-bond interactions, while plasticity in loop 27-36 is essential to bind purine- or pyrimidine-based nucleosides. Further, we deciphered the role of PnrA in pneumococcal fitness in infection experiments. Phagocytosis experiments did not show a clear difference in phagocytosis between PnrA-deficient and wild-type pneumococci. In the acute pneumonia infection model the deficiency of PnrA attenuated moderately virulence of the mutant, which is indicated by a delay in the development of severe lung infections. Importantly, we confirmed the loss of fitness in co-infections, where the wild-type out-competed the pnrA-mutant. In conclusion, we present the PnrA structure in complex with individual nucleosides and show that the consumption of adenosine and guanosine under infection conditions is required for virulence.
Organizational Affiliation:
Department of Molecular Genetics and Infection Biology, Interfaculty Institute for Genetics and Functional Genomics, Center for Functional Genomics of Microbes, University of Greifswald, D-17487 Greifswald, Germany; Present Address: Institut für Klinische Chemie und Laboratoriumsmedizin, Universitätsmedizin Greifswald, Germany.