Optimization of Pyrazoles as Phenol Surrogates to Yield Potent Inhibitors of Macrophage Migration Inhibitory Factor.
Trivedi-Parmar, V., Robertson, M.J., Cisneros, J.A., Krimmer, S.G., Jorgensen, W.L.(2018) ChemMedChem 13: 1092-1097
- PubMed: 29575754
- DOI: https://doi.org/10.1002/cmdc.201800158
- Primary Citation of Related Structures:
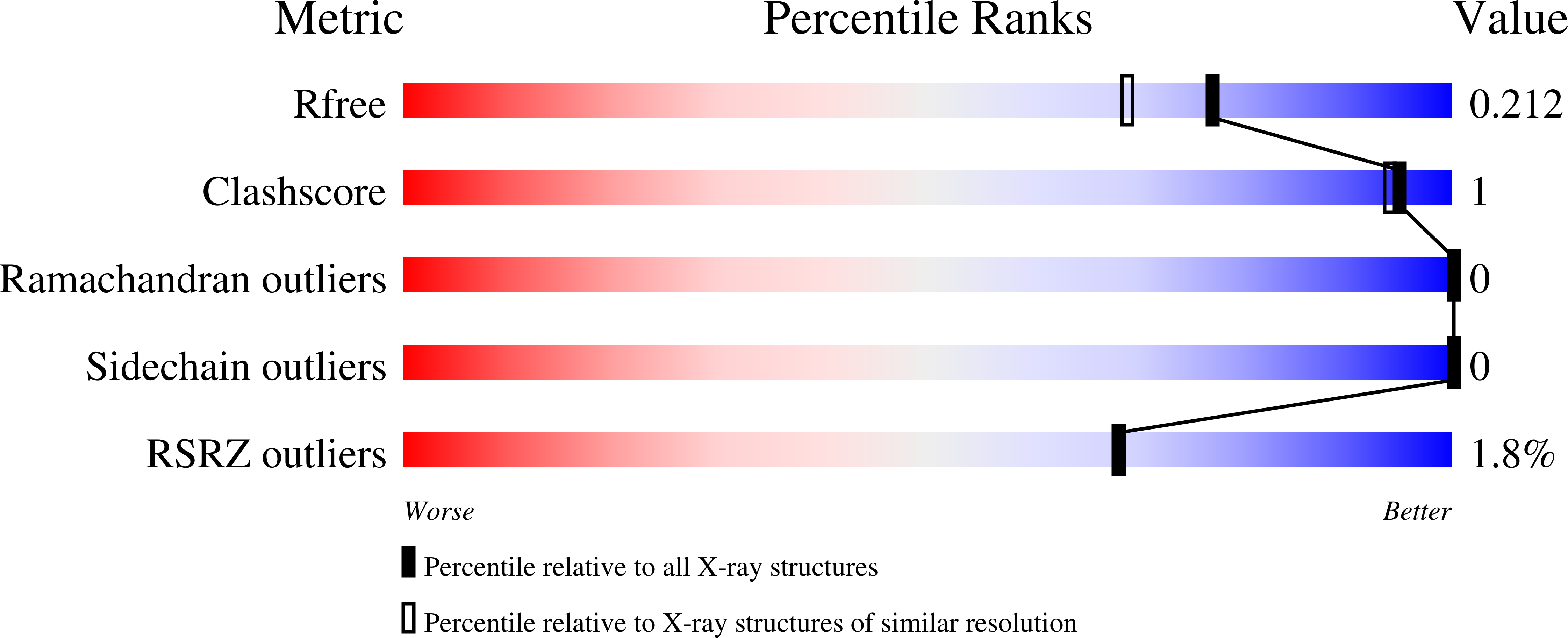
6CB5, 6CBF, 6CBG, 6CBH - PubMed Abstract:
Macrophage migration inhibitory factor (MIF) is a proinflammatory cytokine that is implicated in the regulation of inflammation, cell proliferation, and neurological disorders. MIF is also an enzyme that functions as a keto-enol tautomerase. Most potent MIF tautomerase inhibitors incorporate a phenol, which hydrogen bonds to Asn97 in the active site. Starting from a 113-μm docking hit, we report results of structure-based and computer-aided design that have provided substituted pyrazoles as phenol alternatives with potencies of 60-70 nm. Crystal structures of complexes of MIF with the pyrazoles highlight the contributions of hydrogen bonding with Lys32 and Asn97, and aryl-aryl interactions with Tyr36, Tyr95, and Phe113 to the binding.
Organizational Affiliation:
Department of Chemistry, Yale University, New Haven, CT, 06520-8107, USA.