Optimisation of neuraminidase expression by HEK-293E cells for use in structural biology
Campbell, A.C., Tanner, J.J., Krause, K.L.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Neuraminidase | 404 | Influenza A virus (A/Brevig Mission/1/1918(H1N1)) | Mutation(s): 0 Gene Names: NA EC: 3.2.1.18 | 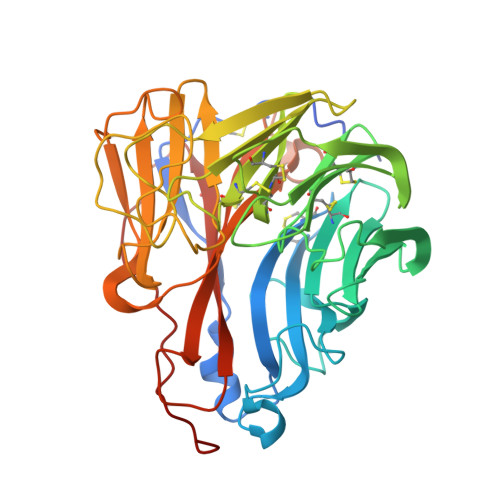 | |
UniProt | |||||
Find proteins for Q9IGQ6 (Influenza A virus (strain A/Brevig Mission/1/1918 H1N1)) Explore Q9IGQ6 Go to UniProtKB: Q9IGQ6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9IGQ6 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AA [auth C] AB [auth H] BA [auth C] FA [auth D] GA [auth D] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| GOL Query on GOL | DB [auth H] EA [auth C] JA [auth D] OA [auth E] U [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CA Query on CA | BB [auth H] CA [auth C] CB [auth H] DA [auth C] HA [auth D] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 122.681 | α = 90 |
| b = 148.189 | β = 94.8 |
| c = 127.166 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| MOLREP | phasing |