Purple acid phosphatase inhibitors as leads for osteoporosis chemotherapeutics.
Hussein, W.M., Feder, D., Schenk, G., Guddat, L.W., McGeary, R.P.(2018) Eur J Med Chem 157: 462-479
- PubMed: 30107365
- DOI: https://doi.org/10.1016/j.ejmech.2018.08.004
- Primary Citation of Related Structures:
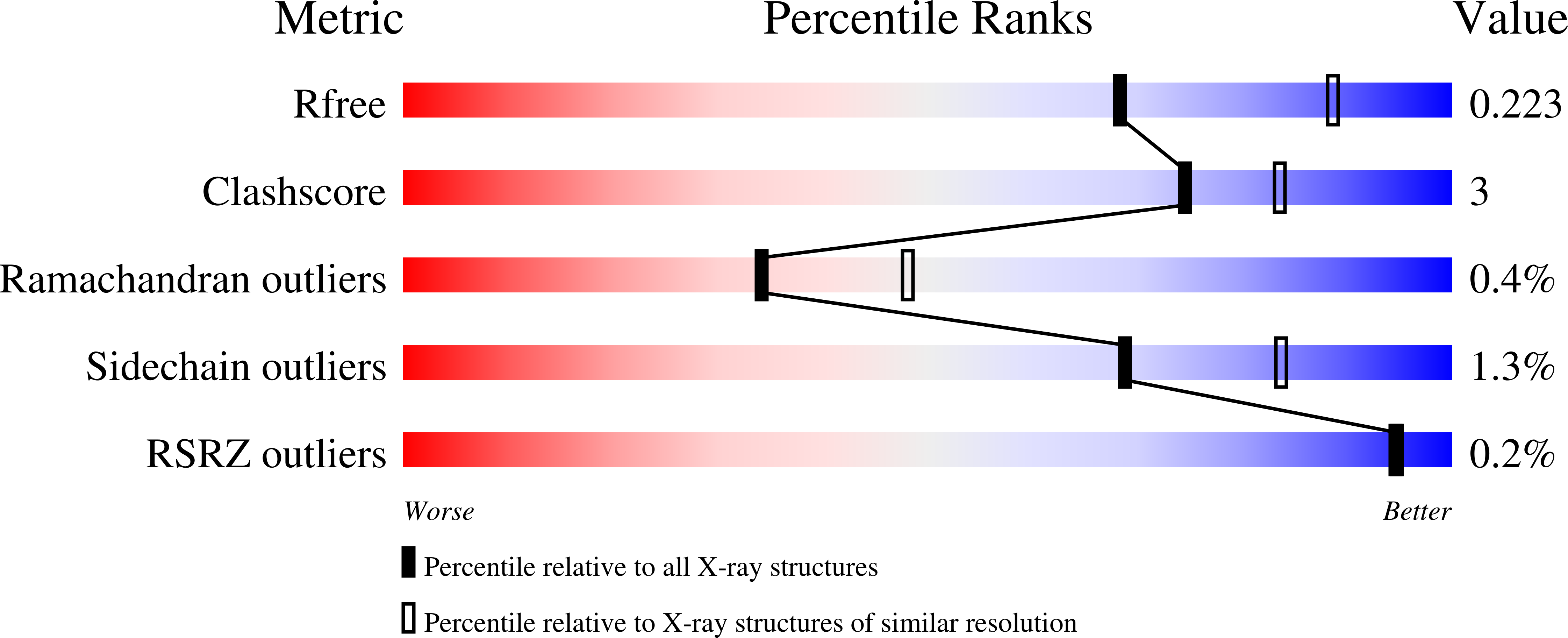
6G46 - PubMed Abstract:
Purple acid phosphatases (PAPs) are metalloenzymes that catalyse the hydrolysis of phosphate esters under acidic conditions. Their active site contains a Fe(III)Fe(II) metal centre in mammals and a Fe(III)Zn(II) or Fe(III)Mn(II) metal centre in plants. In humans, elevated PAP levels in serum strongly correlate with the progression of osteoporosis and metabolic bone malignancies, which make PAP a target suitable for the development of chemotherapeutics to combat bone ailments. Due to difficulties in obtaining the human enzyme, the corresponding enzymes from red kidney bean and pig have been used previously to develop specific PAP inhibitors. Here, existing lead compounds were further elaborated to create a series of inhibitors with K i values as low as ∼30 μM. The inhibition constants of these compounds were of comparable magnitude for pig and red kidney bean PAPs, indicating that relevant binding interactions are conserved. The crystal structure of red kidney bean PAP in complex with the most potent inhibitor in this series, compound 4f, was solved to 2.40 Å resolution. This inhibitor coordinates directly to the binuclear metal centre in the active site as expected based on its competitive mode of inhibition. Docking simulations predict that this compound binds to human PAP in a similar mode. This study presents the first example of a PAP structure in complex with an inhibitor that is of relevance to the development of anti-osteoporotic chemotherapeutics.
Organizational Affiliation:
The University of Queensland, School of Chemistry and Molecular Biosciences, Brisbane, QLD 4072, Australia; Helwan University, Pharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Ein Helwan, Helwan, Egypt.