Structure of human galactokinase in complex with galactose and ADP
Bezerra, G.A., Mackinnon, S., Williams, E., Zhang, M., Arrowsmith, C., Edwards, A., Bountra, C., Lai, K., Yue, W.W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Galactokinase | 399 | Homo sapiens | Mutation(s): 0 Gene Names: GALK1, GALK EC: 2.7.1.6 | 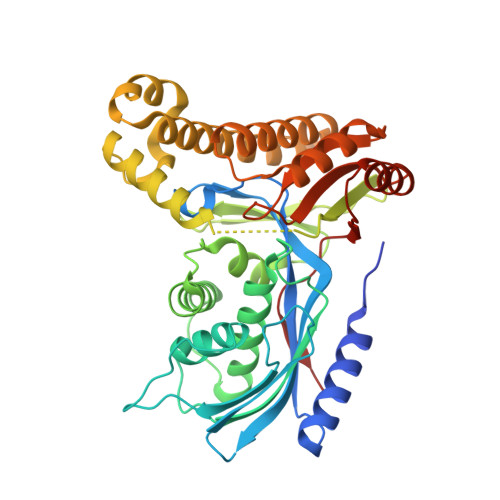 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P51570 (Homo sapiens) Explore P51570 Go to UniProtKB: P51570 | |||||
PHAROS: P51570 GTEx: ENSG00000108479 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P51570 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP Query on ADP | BA [auth H] J [auth A] L [auth B] P [auth C] S [auth D] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| GAL Query on GAL | AA [auth H] I [auth A] K [auth B] O [auth C] R [auth D] | beta-D-galactopyranose C6 H12 O6 WQZGKKKJIJFFOK-FPRJBGLDSA-N |  | ||
| SO4 Query on SO4 | CA [auth H], M [auth B], N [auth B], Q [auth C], X [auth F] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.217 | α = 113.85 |
| b = 110.32 | β = 102.26 |
| c = 118.234 | γ = 101.29 |
| Software Name | Purpose |
|---|---|
| Aimless | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| xia2 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 106169/ZZ14/Z |