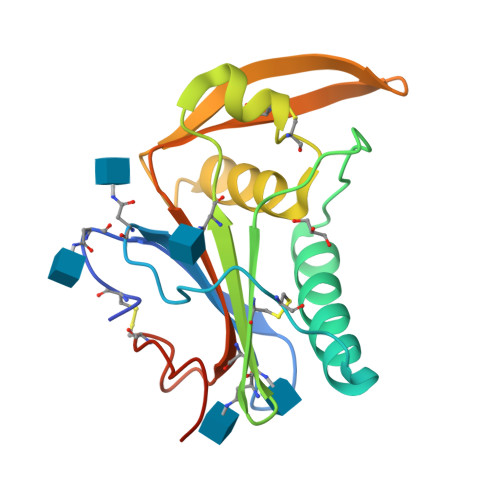
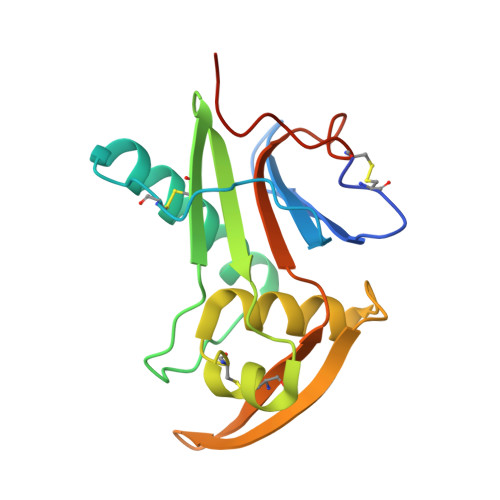
Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
Pryce, R., Ng, W.M., Zeltina, A., Watanabe, Y., El Omari, K., Wagner, A., Bowden, T.A.(2019) J Virol 93
- PubMed: 30305351
- DOI: https://doi.org/10.1128/JVI.01048-18
- Primary Citation of Related Structures:
6HJ4, 6HJ5, 6HJ6, 6HJC - PubMed Abstract:
The emergence of Old and New World arenaviruses from rodent reservoirs persistently threatens human health. The GP1 subunit of the envelope-displayed arenaviral glycoprotein spike complex (GPC) mediates host cell recognition and is an important determinant of cross-species transmission. Previous structural analyses of Old World arenaviral GP1 glycoproteins, alone and in complex with a cognate GP2 subunit, have revealed that GP1 adopts two distinct conformational states distinguished by differences in the orientations of helical regions of the molecule. Here, through comparative study of the GP1 glycoprotein architectures of Old World Loei River virus and New World Whitewater Arroyo virus, we show that these rearrangements are restricted to Old World arenaviruses and are not induced solely by the pH change that is associated with virus endosomal trafficking. Our structure-based phylogenetic analysis of arenaviral GP1s provides a blueprint for understanding the discrete structural classes adopted by these therapeutically important targets. IMPORTANCE The genetically and geographically diverse group of viruses within the family Arenaviridae includes a number of zoonotic pathogens capable of causing fatal hemorrhagic fever. The multisubunit GPC glycoprotein spike complex displayed on the arenavirus envelope is a key determinant of species tropism and a primary target of the host humoral immune response. Here, we show that the receptor-binding GP1 subcomponent of the GPC spike from Old World but not New World arenaviruses adopts a distinct, pH-independent conformation in the absence of the cognate GP2. Our analysis provides a structure-based approach to understanding the discrete conformational classes sampled by these therapeutically important targets, informing strategies to develop arenaviral glycoprotein immunogens that resemble GPC as presented on the mature virion surface.
Organizational Affiliation:
Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, Oxford, United Kingdom.