Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi in complex with guanosine
Volpato Santos, Y., Lorentzen, E., Basquin, J., Boshart, M.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein kinase A regulatory subunit | 304 | Trypanosoma cruzi | Mutation(s): 0 Gene Names: PKAR, C3747_37g237 | 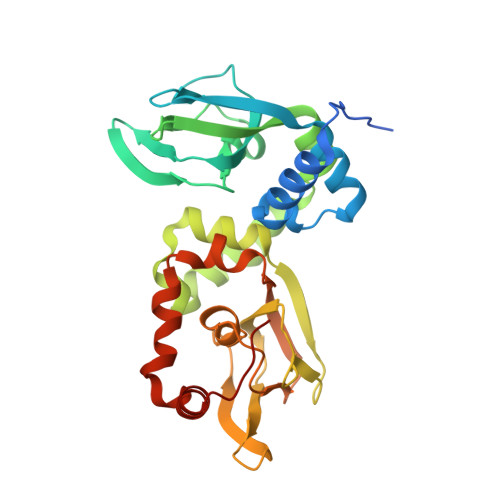 | |
UniProt | |||||
Find proteins for Q86RB0 (Trypanosoma cruzi) Explore Q86RB0 Go to UniProtKB: Q86RB0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q86RB0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GMP (Subject of Investigation/LOI) Query on GMP | E [auth A] F [auth A] G [auth B] H [auth B] J [auth C] | GUANOSINE C10 H13 N5 O5 NYHBQMYGNKIUIF-UUOKFMHZSA-N |  | ||
| ACT Query on ACT | I [auth B] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.91 | α = 90 |
| b = 80.819 | β = 92.799 |
| c = 149.343 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation | Germany | Bo1100/7-1 |