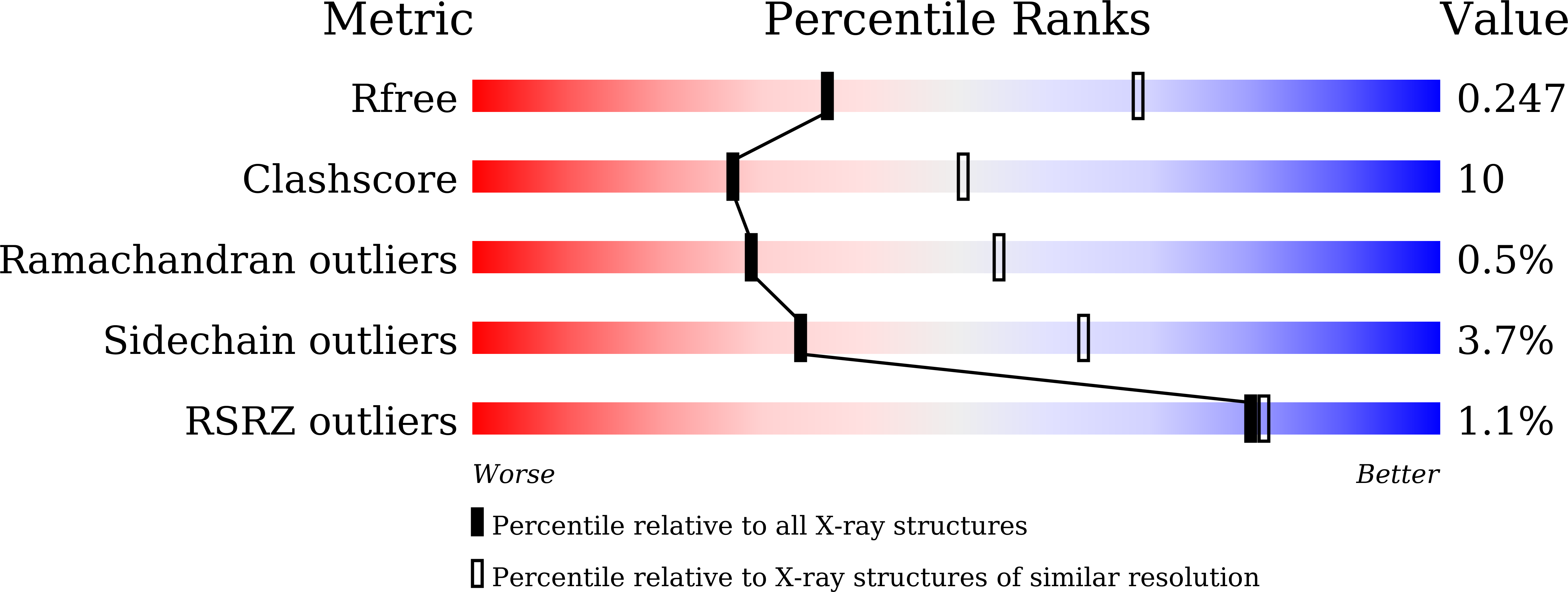
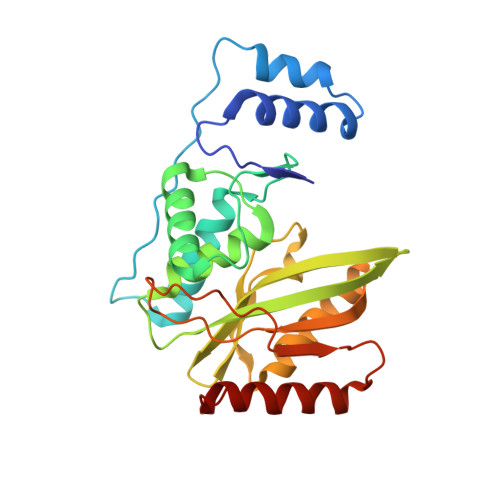
Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Wang, N., Bosc, C., Ryul Choi, S., Boulan, B., Peris, L., Olieric, N., Bao, H., Krichen, F., Chen, L., Andrieux, A., Olieric, V., Moutin, M.J., Steinmetz, M.O., Huang, H.(2019) Nat Struct Mol Biol 26: 571-582
- PubMed: 31235911
- DOI: https://doi.org/10.1038/s41594-019-0241-y
- Primary Citation of Related Structures:
6J4O, 6J4P, 6J4Q, 6J4S, 6J4U, 6J4V, 6QBY - PubMed Abstract:
Vasohibins are tubulin tyrosine carboxypeptidases that are important in neuron physiology. We examined the crystal structures of human vasohibin 1 and 2 in complex with small vasohibin-binding protein (SVBP) in the absence and presence of different inhibitors and a C-terminal α-tubulin peptide. In combination with functional data, we propose that SVBP acts as an activator of vasohibins. An extended groove and a distinctive surface residue patch of vasohibins define the specific determinants for recognizing and cleaving the C-terminal tyrosine of α-tubulin and for binding microtubules, respectively. The vasohibin-SVBP interaction and the ability of the enzyme complex to associate with microtubules regulate axon specification of neurons. Our results define the structural basis of tubulin detyrosination by vasohibins and show the relevance of this process for neuronal development. Our findings offer a unique platform for developing drugs against human conditions with abnormal tubulin tyrosination levels, such as cancer, heart defects and possibly brain disorders.
Organizational Affiliation:
Department of Biology, Southern University of Science and Technology, Shenzhen, China.