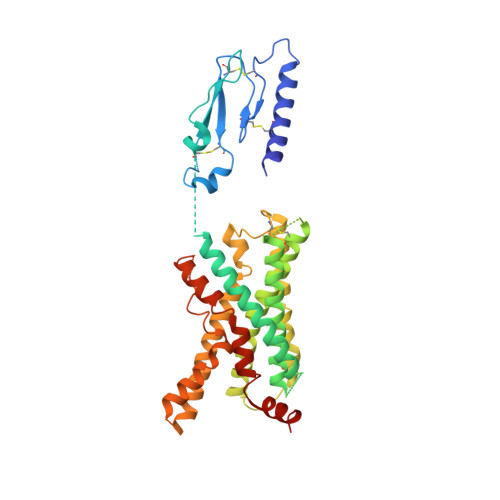
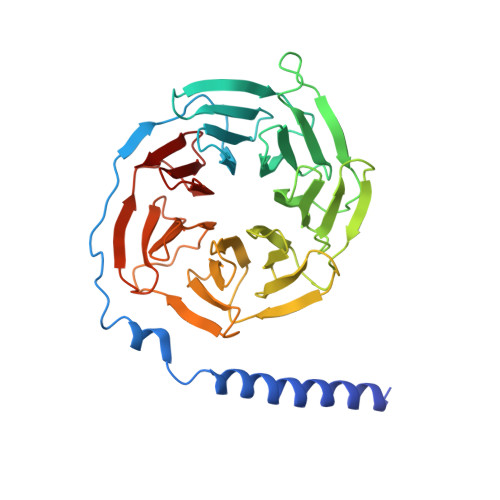
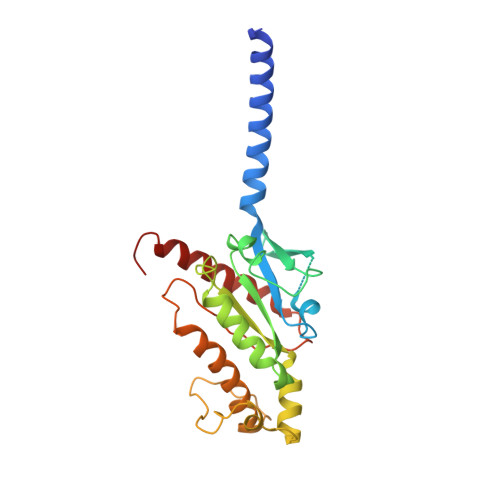
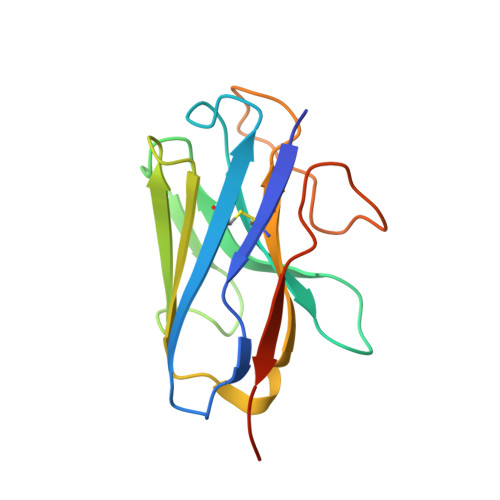
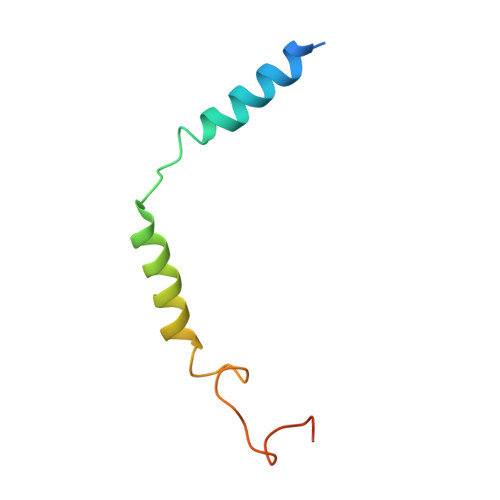
Cryo-EM structure of the human PAC1 receptor coupled to an engineered heterotrimeric G protein.
Kobayashi, K., Shihoya, W., Nishizawa, T., Kadji, F.M.N., Aoki, J., Inoue, A., Nureki, O.(2020) Nat Struct Mol Biol 27: 274-280
- PubMed: 32157248
- DOI: https://doi.org/10.1038/s41594-020-0386-8
- Primary Citation of Related Structures:
6LPB - PubMed Abstract:
Pituitary adenylate cyclase-activating polypeptide (PACAP) is a pleiotropic neuropeptide hormone. The PACAP receptor PAC1R, which belongs to the class B G-protein-coupled receptors (GPCRs), is a drug target for mental disorders and dry eye syndrome. Here, we present a cryo-EM structure of human PAC1R bound to PACAP and an engineered G s heterotrimer. The structure revealed that transmembrane helix TM1 plays an essential role in PACAP recognition. The extracellular domain (ECD) of PAC1R tilts by ~40° compared with that of the glucagon-like peptide-1 receptor (GLP-1R) and thus does not cover the peptide ligand. A functional analysis demonstrated that the PAC1R ECD functions as an affinity trap and is not required for receptor activation, whereas the GLP-1R ECD plays an indispensable role in receptor activation, illuminating the functional diversity of the ECDs in class B GPCRs. Our structural information will facilitate the design and improvement of better PAC1R agonists for clinical applications.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo, Tokyo, Japan.