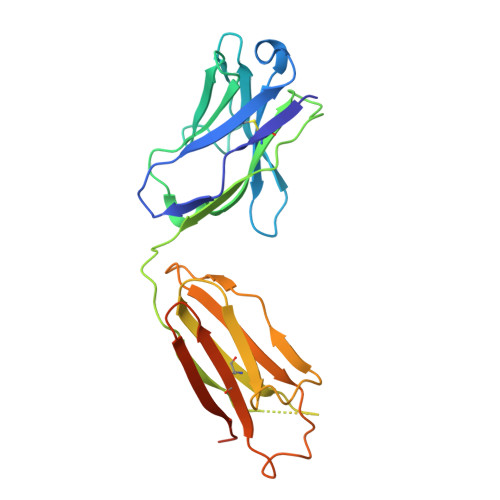
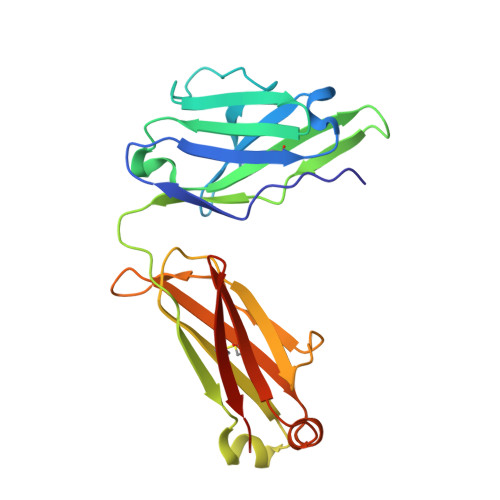
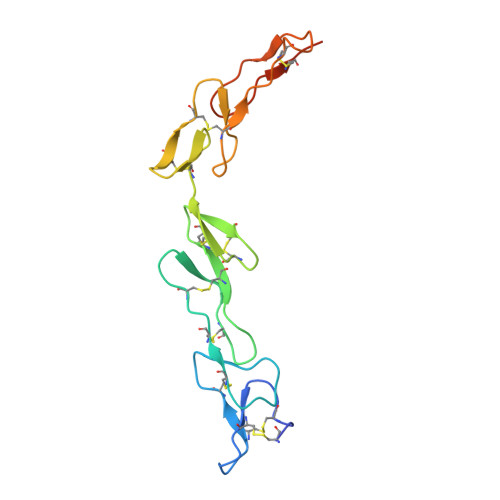
Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Chin, S.M., Kimberlin, C.R., Roe-Zurz, Z., Zhang, P., Xu, A., Liao-Chan, S., Sen, D., Nager, A.R., Oakdale, N.S., Brown, C., Wang, F., Yang, Y., Lindquist, K., Yeung, Y.A., Salek-Ardakani, S., Chaparro-Riggers, J.(2018) Nat Commun 9: 4679-4679
- PubMed: 30410017
- DOI: https://doi.org/10.1038/s41467-018-07136-7
- Primary Citation of Related Structures:
6MGE, 6MHR, 6MI2 - PubMed Abstract:
4-1BB (CD137, TNFRSF9) is an inducible costimulatory receptor expressed on activated T cells. Clinical trials of two agonist antibodies, utomilumab (PF-05082566) and urelumab (BMS-663513), are ongoing in multiple cancer indications, and both antibodies demonstrate distinct activities in the clinic. To understand these differences, we solved structures of the human 4-1BB/4-1BBL complex, the 4-1BBL trimer alone, and 4-1BB bound to utomilumab or urelumab. The 4-1BB/4-1BBL complex displays a unique interaction between receptor and ligand when compared with other TNF family members. Furthermore, our ligand-only structure differs from previously published data. Utomilumab, a ligand-blocking antibody, binds 4-1BB between CRDs 3 and 4. In contrast, urelumab binds 4-1BB CRD-1, away from the ligand binding site. Finally, cell-based assays demonstrate utomilumab is a milder agonist than urelumab. Collectively, our data provide a deeper understanding of the 4-1BB signaling complex, providing a template for future development of next generation 4-1BB targeted biologics.
Organizational Affiliation:
Cancer Immunology Discovery, Pfizer Inc., 230 E. Grand Ave, South San Francisco, CA, 94080, USA. shermanmichael.chin@pfizer.com.