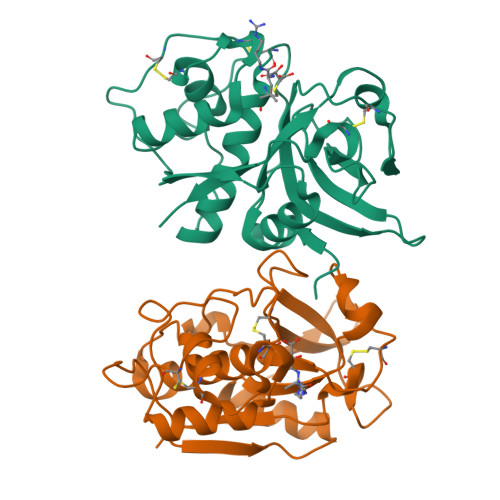
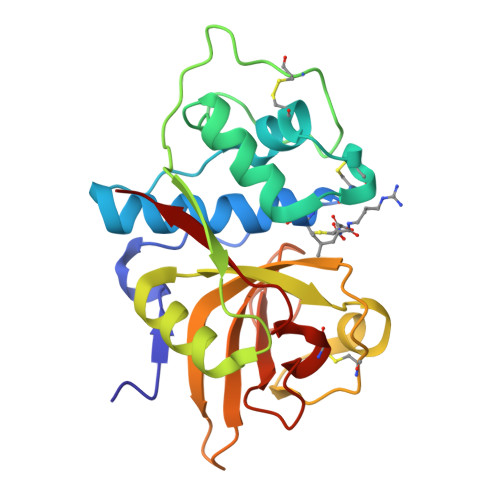
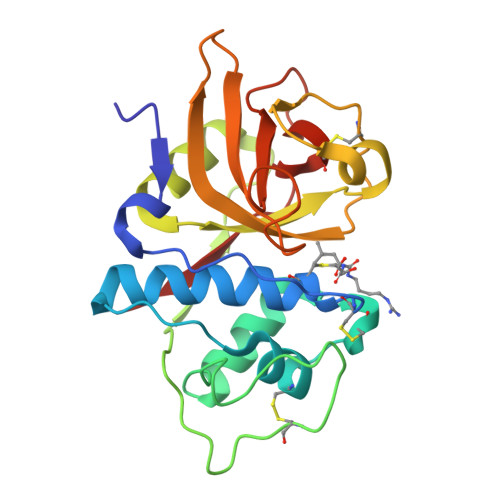
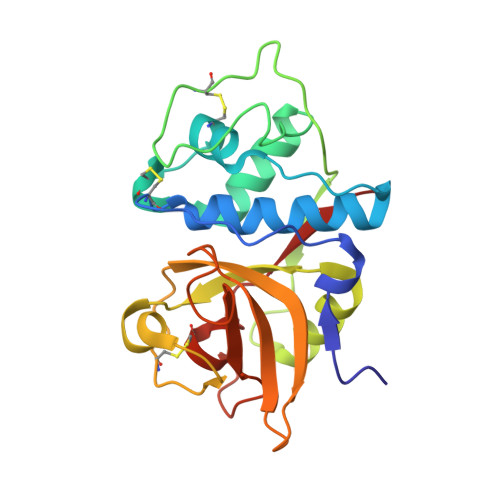
Determination of the crystal structure and substrate specificity of ananain.
Yongqing, T., Wilmann, P.G., Pan, J., West, M.L., Brown, T.J., Mynott, T., Pike, R.N., Wijeyewickrema, L.C.(2019) Biochimie 166: 194-202
- PubMed: 31306685
- DOI: https://doi.org/10.1016/j.biochi.2019.07.011
- Primary Citation of Related Structures:
6MIS, 6OKJ - PubMed Abstract:
Ananain (EC 3.4.22.31) accounts for less than 10% of the total enzyme in the crude pineapple stem extract known as bromelain, yet yields the majority of the proteolytic activity of bromelain. Despite a high degree of sequence identity between ananain and stem bromelain, the most abundant bromelain cysteine protease, ananain displays distinct chemical properties, substrate preference and inhibitory profile compared to stem bromelain. A tripeptidyl substrate library (REPLi) was used to further characterize the substrate specificity of ananain and identified an optimal substrate for cleavage by ananain. The optimal tripeptide, PLQ, yielded a high k cat /K m value of 1.7 x 106 M -1 s -1 , with cleavage confirmed to occur after the Gln residue. Crystal structures of unbound ananain and an inhibitory complex of ananain and E-64, solved at 1.73 and 1.98 Å, respectively, revealed a geometrically flat and open S1 subsite for ananain. This subsite accommodates diverse P1 substrate residues, while a narrow and deep hydrophobic pocket-like S2 subsite would accommodate a non-polar P2 residue, such as the preferred Leu residue observed in the specificity studies. A further illustration of the atomic interactions between E-64 and ananain explains the high inhibitory efficiency of E-64 toward ananain. These data reveal the first in depth structural and functional data for ananain and provide a basis for further study of the natural properties of the enzyme.
Organizational Affiliation:
Department of Biochemistry & Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Australia; Anatara Lifesciences Ltd., Brisbane, Australia.