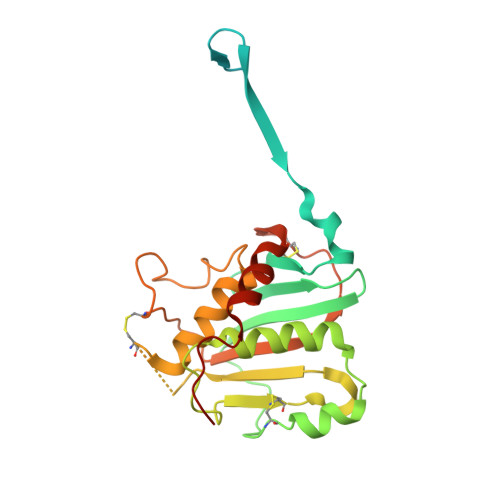
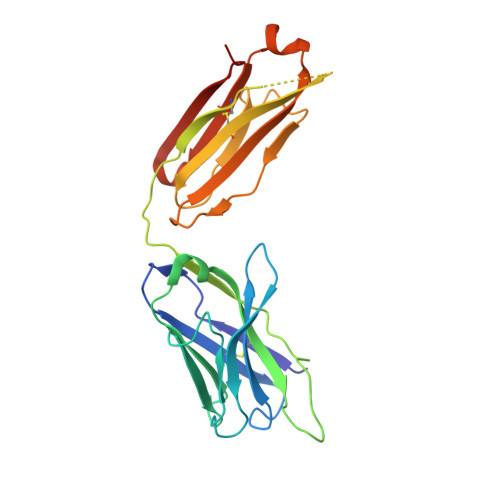
Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Hastie, K.M., Cross, R.W., Harkins, S.S., Zandonatti, M.A., Koval, A.P., Heinrich, M.L., Rowland, M.M., Robinson, J.E., Geisbert, T.W., Garry, R.F., Branco, L.M., Saphire, E.O.(2019) Cell 178: 1004-1015.e14
- PubMed: 31398326
- DOI: https://doi.org/10.1016/j.cell.2019.07.020
- Primary Citation of Related Structures:
6P91, 6P95 - PubMed Abstract:
Lassa virus (LASV) causes hemorrhagic fever and is endemic in West Africa. Protective antibody responses primarily target the LASV surface glycoprotein (GPC), and GPC-B competition group antibodies often show potent neutralizing activity in humans. However, which features confer potent and broadly neutralizing antibody responses is unclear. Here, we compared three crystal structures of LASV GPC complexed with GPC-B antibodies of varying neutralization potency. Each GPC-B antibody recognized an overlapping epitope involved in binding of two adjacent GPC monomers and preserved the prefusion trimeric conformation. Differences among GPC-antibody interactions highlighted specific residues that enhance neutralization. Using structure-guided amino acid substitutions, we increased the neutralization potency and breadth of these antibodies to include all major LASV lineages. The ability to define antibody residues that allow potent and broad neutralizing activity, together with findings from analyses of inferred germline precursors, is critical to develop potent therapeutics and for vaccine design and assessment.
Organizational Affiliation:
La Jolla Institute for Immunology, La Jolla, CA, USA; Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA, USA.