Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Shaw, S.A., Vokits, B.P., Dilger, A.K., Viet, A., Clark, C.G., Abell, L.M., Locke, G.A., Duke, G., Kopcho, L.M., Dongre, A., Gao, J., Krishnakumar, A., Jusuf, S., Khan, J., Spronk, S.A., Basso, M.D., Zhao, L., Cantor, G.H., Onorato, J.M., Wexler, R.R., Duclos, F., Kick, E.K.(2020) Bioorg Med Chem 28: 115723-115723
- PubMed: 33007547
- DOI: https://doi.org/10.1016/j.bmc.2020.115723
- Primary Citation of Related Structures:
6WXZ, 6WY0, 6WY5, 6WY7, 6WYD - PubMed Abstract:
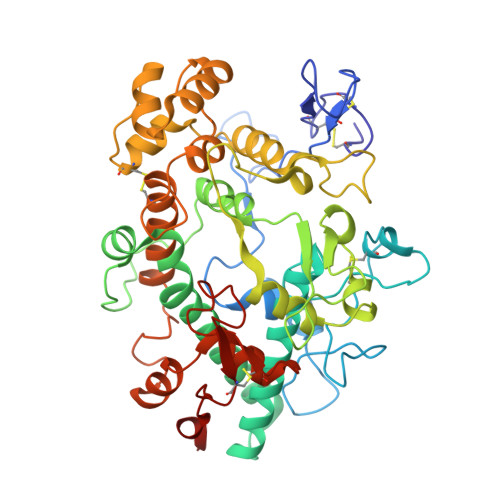
Myeloperoxidase (MPO) is a heme peroxidase found in neutrophils, monocytes and macrophages that efficiently catalyzes the oxidation of endogenous chloride into hypochlorous acid for antimicrobial activity. Chronic MPO activation can lead to indiscriminate protein modification causing tissue damage, and has been associated with chronic inflammatory diseases, atherosclerosis, and acute cardiovascular events. Triazolopyrimidine 5 is a reversible MPO inhibitor; however it suffers from poor stability in acid, and is an irreversible inhibitor of the DNA repair protein methyl guanine methyl transferase (MGMT). Structure-based drug design was employed to discover benzyl triazolopyridines with improved MPO potency, as well as acid stability, no reactivity with MGMT, and selectivity against thyroid peroxidase (TPO). Structure-activity relationships, a crystal structure of the MPO-inhibitor complex, and acute in vivo pharmacodynamic data are described herein.
Organizational Affiliation:
Bristol Myers Squibb Company, P.O. Box 5400, Princeton, NJ 08543-5400, United States. Electronic address: scott.shaw@bms.com.