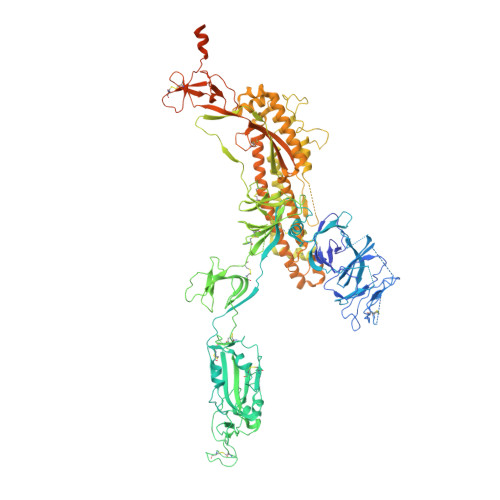
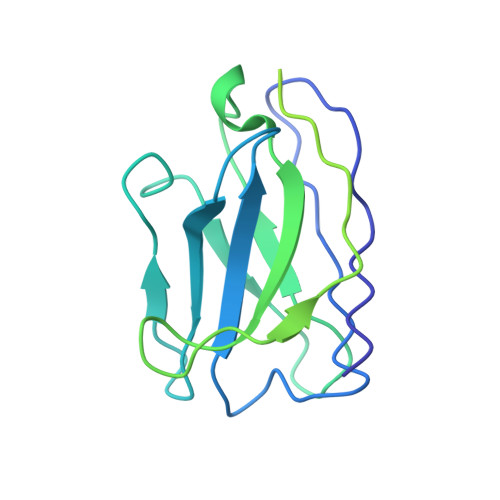
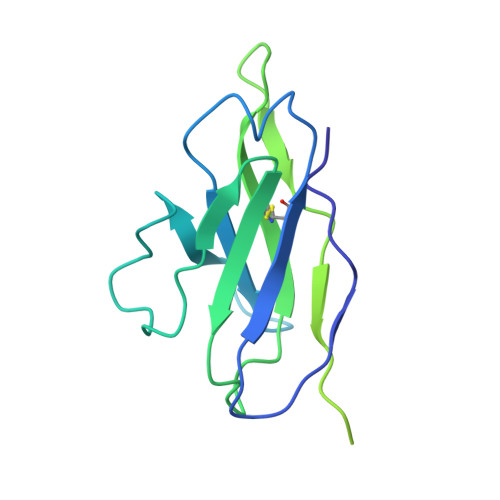
Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies.
Barnes, C.O., West Jr., A.P., Huey-Tubman, K.E., Hoffmann, M.A.G., Sharaf, N.G., Hoffman, P.R., Koranda, N., Gristick, H.B., Gaebler, C., Muecksch, F., Lorenzi, J.C.C., Finkin, S., Hagglof, T., Hurley, A., Millard, K.G., Weisblum, Y., Schmidt, F., Hatziioannou, T., Bieniasz, P.D., Caskey, M., Robbiani, D.F., Nussenzweig, M.C., Bjorkman, P.J.(2020) Cell 182: 828
- PubMed: 32645326
- DOI: https://doi.org/10.1016/j.cell.2020.06.025
- Primary Citation of Related Structures:
6XCA, 6XCM, 6XCN - PubMed Abstract:
Neutralizing antibody responses to coronaviruses mainly target the receptor-binding domain (RBD) of the trimeric spike. Here, we characterized polyclonal immunoglobulin Gs (IgGs) and Fabs from COVID-19 convalescent individuals for recognition of coronavirus spikes. Plasma IgGs differed in their focus on RBD epitopes, recognition of alpha- and beta-coronaviruses, and contributions of avidity to increased binding/neutralization of IgGs over Fabs. Using electron microscopy, we examined specificities of polyclonal plasma Fabs, revealing recognition of both S1 A and RBD epitopes on SARS-CoV-2 spike. Moreover, a 3.4 Å cryo-electron microscopy (cryo-EM) structure of a neutralizing monoclonal Fab-spike complex revealed an epitope that blocks ACE2 receptor binding. Modeling based on these structures suggested different potentials for inter-spike crosslinking by IgGs on viruses, and characterized IgGs would not be affected by identified SARS-CoV-2 spike mutations. Overall, our studies structurally define a recurrent anti-SARS-CoV-2 antibody class derived from VH3-53/VH3-66 and similarity to a SARS-CoV VH3-30 antibody, providing criteria for evaluating vaccine-elicited antibodies.
Organizational Affiliation:
Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA, USA.