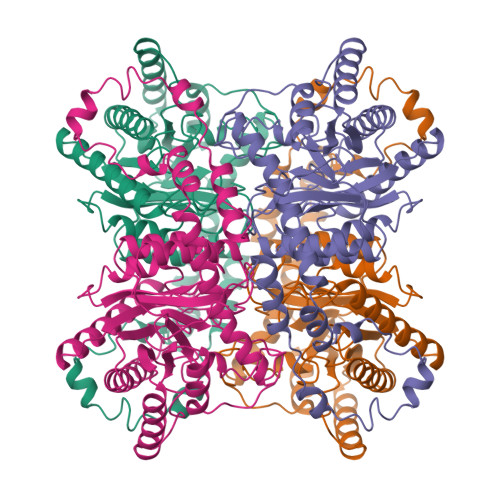
Refinement of glucose isomerase from Streptomyces albus at 1.65 A with data from an imaging plate.
Dauter, Z., Terry, H., Witzel, H., Wilson, K.S.(1990) Acta Crystallogr B 46: 833-841
- PubMed: 2085424
- DOI: https://doi.org/10.1107/s0108768190008059
- Primary Citation of Related Structures:
6XIA - PubMed Abstract:
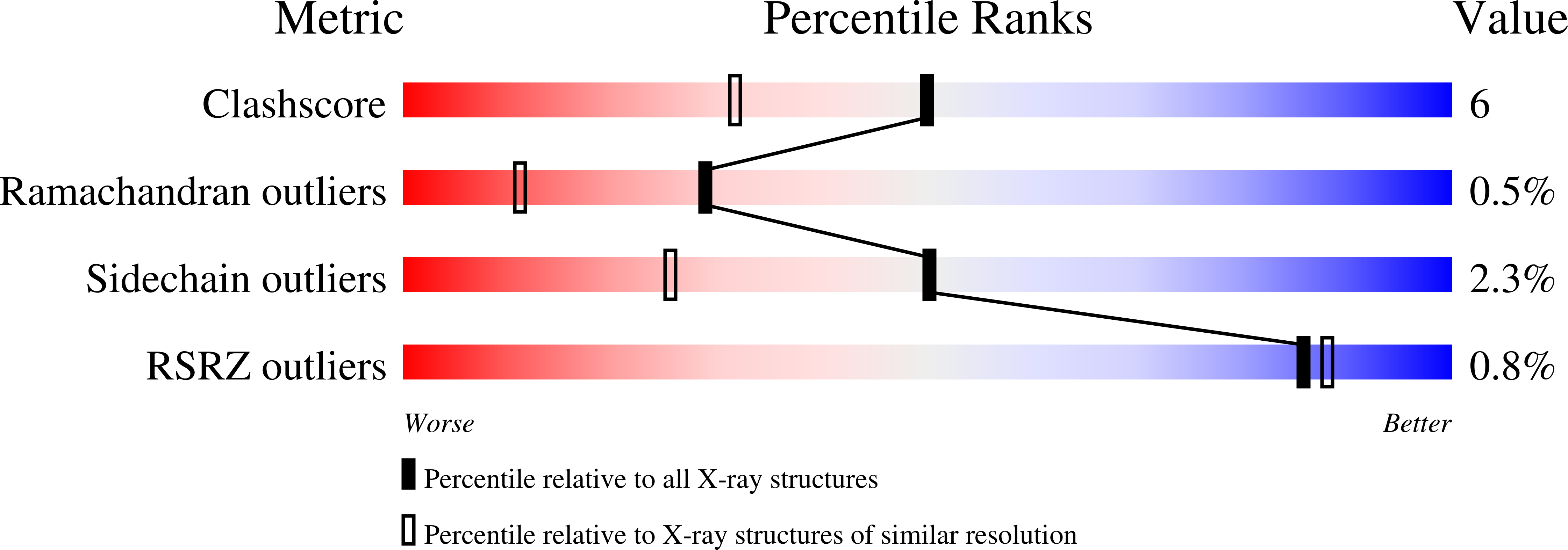
The structure of 'metal-free' glucose isomerase of Streptomyces albus strain number YT ATCC 21132 has been analysed and refined at 1.65 A. The space group is I222, with cell dimensions a = 93.9 (1), b = 99.7 (1) and c = 102.9 (1) A, and there is one monomer of the tetrameric molecule per asymmetric unit. The data were recorded from two crystals of the protein using synchrotron radiation from the EMBL beamline X11 at DESY, Hamburg. Data were recorded with an imaging plate scanner designed and built in the EMBL Hamburg outstation. The total data-collection time was less than 12 h and the processing of all data took less than 2 days. The coordinates of the Arthrobacter glucose isomerase refined at a resolution of 2.5 A were used as a starting model. The structure of the protein and of 445 associated water molecules in the asymmetric unit were refined by restrained least-squares minimization using all data between 8 and 1.65 A to a final R factor of 14.1%.
Organizational Affiliation:
European Molecular Biology Laboratory (EMBL), c/o DESY, Hamburg, Federal Republic of Germany.