Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Qiang, M., Ma, P., Li, Y., Liu, H., Harding, A., Min, C., Wang, F., Liu, L., Yuan, M., Ji, Q., Tao, P., Shi, X., Li, Z., Li, T., Wang, X., Zhang, Y., Wu, N.C., Lee, C.D., Zhu, X., Gilbert-Jaramillo, J., Zhang, C., Saxena, A., Huang, X., Wang, H., James, W., Dwek, R.A., Wilson, I.A., Yang, G., Lerner, R.A.(2022) Adv Sci (Weinh) 9: e2102181-e2102181
- PubMed: 34716683
- DOI: https://doi.org/10.1002/advs.202102181
- Primary Citation of Related Structures:
7KN3, 7KN4 - PubMed Abstract:
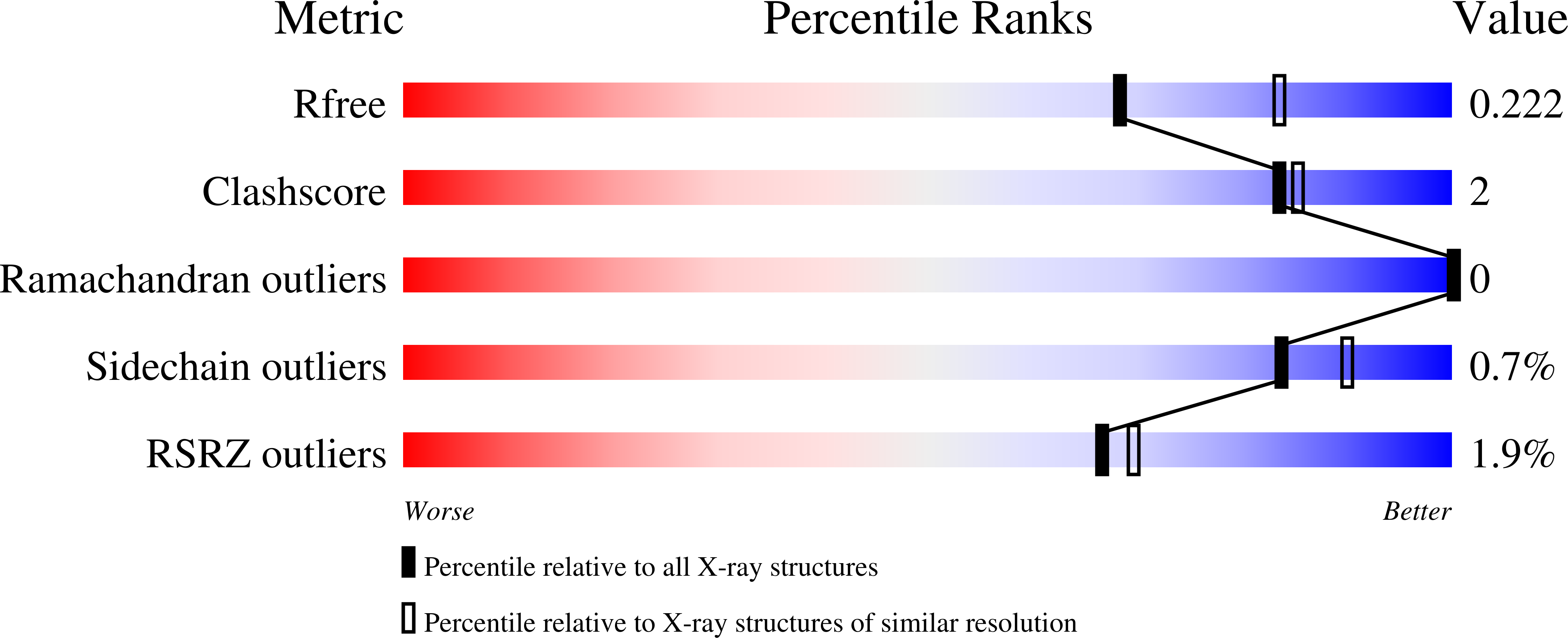
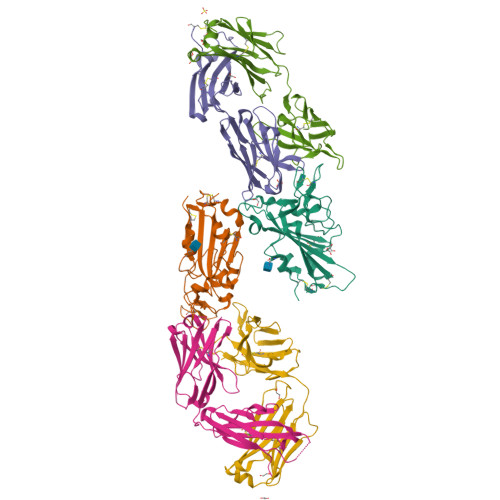
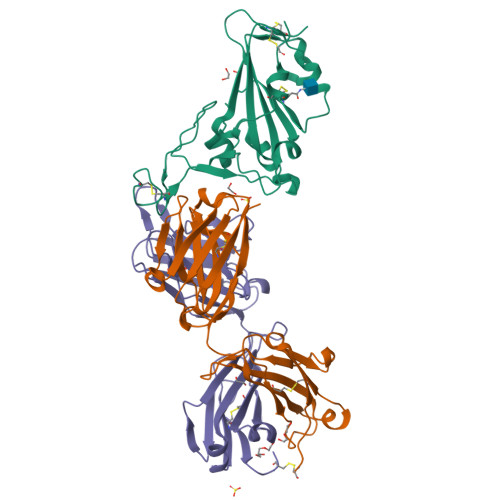
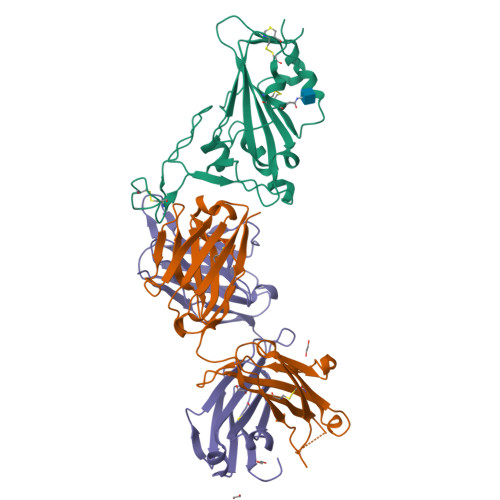
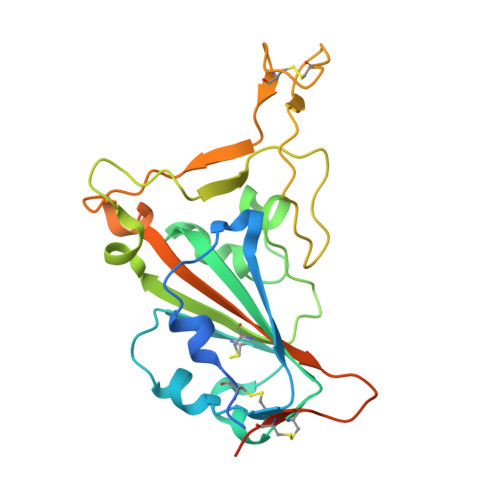
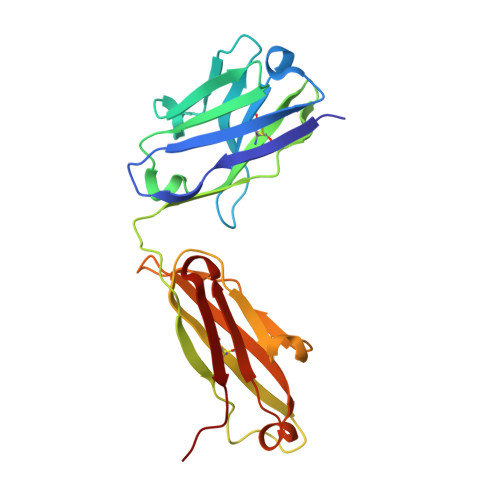
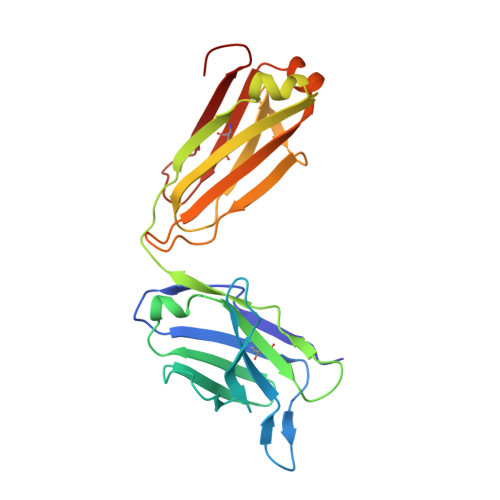
Combinatorial antibody libraries not only effectively reduce antibody discovery to a numbers game, but enable documentation of the history of antibody responses in an individual. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has prompted a wider application of this technology to meet the public health challenge of pandemic threats in the modern era. Herein, a combinatorial human antibody library constructed 20 years before the coronavirus disease 2019 (COVID-19) pandemic is used to discover three highly potent antibodies that selectively bind SARS-CoV-2 spike protein and neutralize authentic SARS-CoV-2 virus. Compared to neutralizing antibodies from COVID-19 patients with generally low somatic hypermutation (SHM), these three antibodies contain over 13-22 SHMs, many of which are involved in specific interactions in their crystal structures with SARS-CoV-2 spike receptor binding domain. The identification of these somatically mutated antibodies in a pre-pandemic library raises intriguing questions about the origin and evolution of these antibodies with respect to their reactivity with SARS-CoV-2.
Organizational Affiliation:
Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai, 201210, P. R. China.