Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Tong, P., Gautam, A., Windsor, I., Travers, M., Chen, Y., Garcia, N., Whiteman, N.B., McKay, L.G.A., Lelis, F.J.N., Habibi, S., Cai, Y., Rennick, L.J., Duprex, W.P., McCarthy, K.R., Lavine, C.L., Zuo, T., Lin, J., Zuiani, A., Feldman, J., MacDonald, E.A., Hauser, B.M., Griffths, A., Seaman, M.S., Schmidt, A.G., Chen, B., Neuberg, D., Bajic, G., Harrison, S.C., Wesemann, D.R.(2021) bioRxiv
- PubMed: 33758863
- DOI: https://doi.org/10.1101/2021.03.10.434840
- Primary Citation of Related Structures:
7N62, 7N64 - PubMed Abstract:
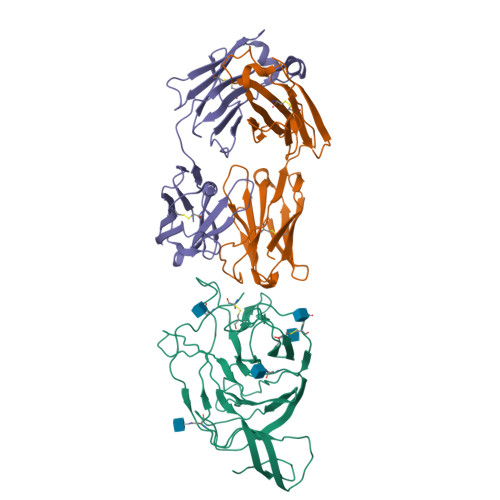
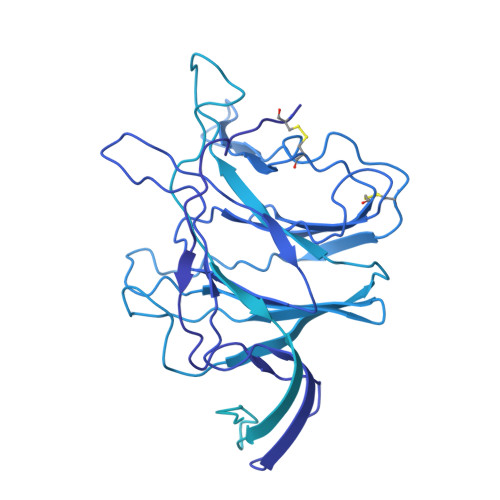
Memory B cell reserves can generate protective antibodies against repeated SARS-CoV-2 infections, but with an unknown reach from original infection to antigenically drifted variants. We charted memory B cell receptor-encoded monoclonal antibodies (mAbs) from 19 COVID-19 convalescent subjects against SARS-CoV-2 spike (S) and found 7 major mAb competition groups against epitopes recurrently targeted across individuals. Inclusion of published and newly determined structures of mAb-S complexes identified corresponding epitopic regions. Group assignment correlated with cross-CoV-reactivity breadth, neutralization potency, and convergent antibody signatures. mAbs that competed for binding the original S isolate bound differentially to S variants, suggesting the protective importance of otherwise-redundant recognition. The results furnish a global atlas of the S-specific memory B cell repertoire and illustrate properties conferring robustness against emerging SARS-CoV-2 variants.
Organizational Affiliation:
Department of Medicine, Division of Allergy and Immunology, Division of Genetics, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.