Structural and Functional Characterization of Camelus dromedarius Glutathione Transferase M1-1.
Perperopoulou, F., Poudel, N., Papageorgiou, A.C., Ataya, F.S., Labrou, N.E.(2022) Life (Basel) 12
- PubMed: 35054499
- DOI: https://doi.org/10.3390/life12010106
- Primary Citation of Related Structures:
7OPY, 7OPZ - PubMed Abstract:
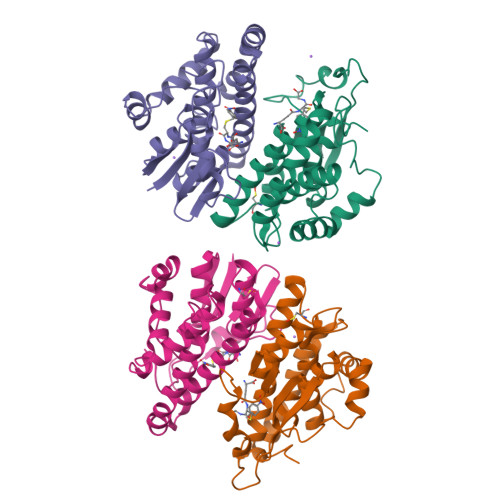
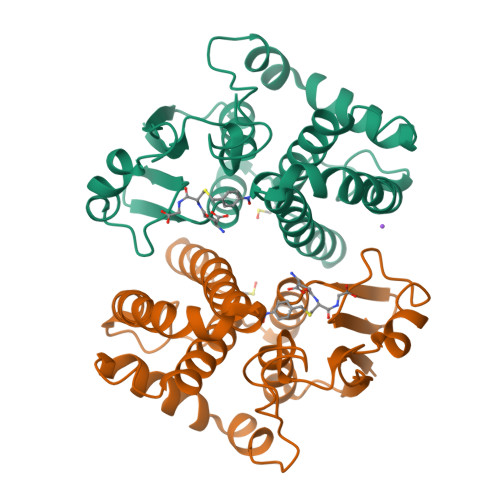
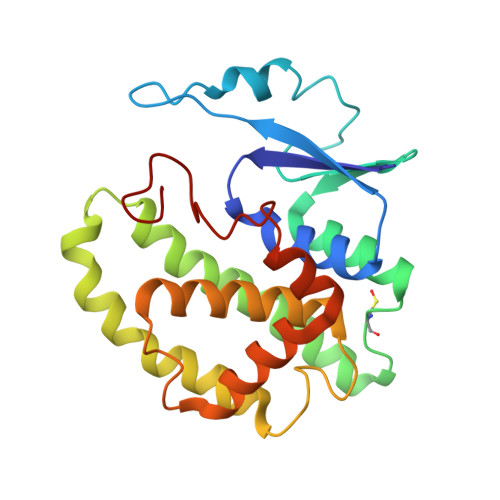
Glutathione transferases (GSTs; EC. 2.5.1.18) are a large family of multifunctional enzymes that play crucial roles in the metabolism and inactivation of a broad range of xenobiotic compounds. In the present work, we report the kinetic and structural characterization of the isoenzyme GSTM1-1 from Camelus dromedarius ( Cd GSTM1-1). The Cd GSΤM1-1 was expressed in E. coli BL21 (DE3) and was purified by affinity chromatography. Kinetics analysis showed that the enzyme displays a relative narrow substrate specificity and restricted ability to bind xenobiotic compounds. The crystal structures of Cd GSΤM1-1 were determined by X-ray crystallography in complex with the substrate (GSH) or the reaction product (S-p-nitrobenzyl-GSH), providing snapshots of the induced-fit catalytic mechanism. The thermodynamic stability of Cd GSTM1-1 was investigated using differential scanning fluorimetry (DSF) in the absence and in presence of GSH and S-p-nitrobenzyl-GSH and revealed that the enzyme's structure is significantly stabilized by its ligands. The results of the present study advance the understanding of camelid GST detoxification mechanisms and their contribution to abiotic stress adaptation in harsh desert conditions.
Organizational Affiliation:
Laboratory of Enzyme Technology, Department of Biotechnology, School of Applied Biology and Biotechnology, Agricultural University of Athens, 75 Iera Odos Street, GR-11855 Athens, Greece.