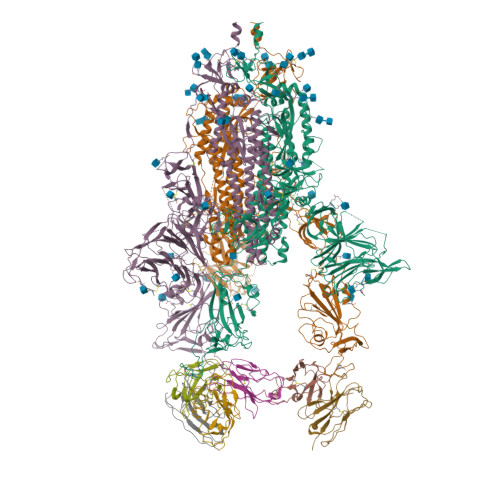
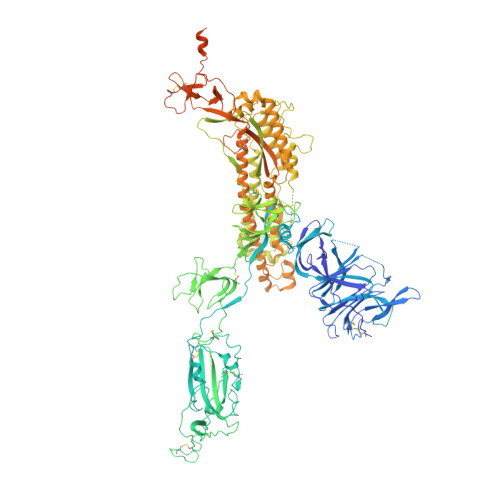
An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern.
Du, W., Hurdiss, D.L., Drabek, D., Mykytyn, A.Z., Kaiser, F.K., Gonzalez-Hernandez, M., Munoz-Santos, D., Lamers, M.M., van Haperen, R., Li, W., Drulyte, I., Wang, C., Sola, I., Armando, F., Beythien, G., Ciurkiewicz, M., Baumgartner, W., Guilfoyle, K., Smits, T., van der Lee, J., van Kuppeveld, F.J.M., van Amerongen, G., Haagmans, B.L., Enjuanes, L., Osterhaus, A.D.M.E., Grosveld, F., Bosch, B.J.(2022) Sci Immunol 7: eabp9312-eabp9312
- PubMed: 35471062
- DOI: https://doi.org/10.1126/sciimmunol.abp9312
- Primary Citation of Related Structures:
7R40 - PubMed Abstract:
The ongoing evolution of SARS-CoV-2 has resulted in the emergence of Omicron, which displays notable immune escape potential through mutations at key antigenic sites on the spike protein. Many of these mutations localize to the spike protein ACE2 receptor binding domain, annulling the neutralizing activity of therapeutic antibodies that were effective against other variants of concern (VOCs) earlier in the pandemic. Here, we identified a receptor-blocking human monoclonal antibody, 87G7, that retained potent in vitro neutralizing activity against SARS-CoV-2 variants including the Alpha, Beta, Gamma, Delta, and Omicron (BA.1/BA.2) VOCs. Using cryo-electron microscopy and site-directed mutagenesis experiments, we showed that 87G7 targets a patch of hydrophobic residues in the ACE2-binding site that are highly conserved in SARS-CoV-2 variants, explaining its broad neutralization capacity. 87G7 protected mice and hamsters prophylactically against challenge with all current SARS-CoV-2 VOCs and showed therapeutic activity against SARS-CoV-2 challenge in both animal models. Our findings demonstrate that 87G7 holds promise as a prophylactic or therapeutic agent for COVID-19 that is more resilient to SARS-CoV-2 antigenic diversity.
Organizational Affiliation:
Virology Section, Infectious Diseases and Immunology Division, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, Netherlands.