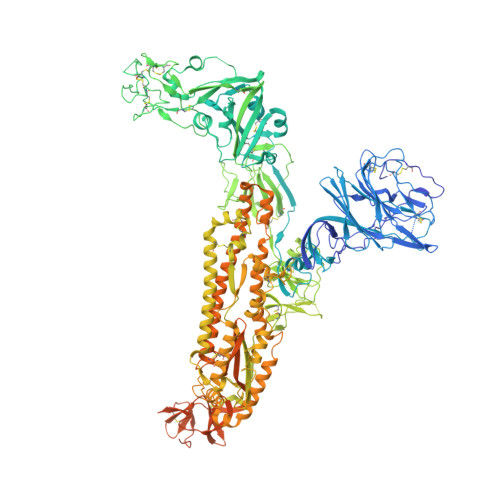
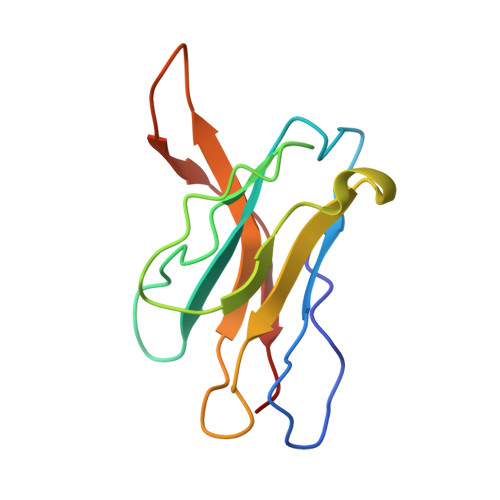
Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Bangaru, S., Antanasijevic, A., Kose, N., Sewall, L.M., Jackson, A.M., Suryadevara, N., Zhan, X., Torres, J.L., Copps, J., de la Pena, A.T., Crowe Jr., J.E., Ward, A.B.(2022) Sci Adv 8: eabn2911-eabn2911
- PubMed: 35507649
- DOI: https://doi.org/10.1126/sciadv.abn2911
- Primary Citation of Related Structures:
7SB3, 7SB4, 7SB5, 7SBV, 7SBW, 7SBX, 7SBY - PubMed Abstract:
Preexisting immunity against seasonal coronaviruses (CoVs) represents an important variable in predicting antibody responses and disease severity to severe acute respiratory syndrome CoV-2 (SARS-CoV-2) infections. We used electron microscopy-based polyclonal epitope mapping (EMPEM) to characterize the antibody specificities against β-CoV spike proteins in prepandemic (PP) sera or SARS-CoV-2 convalescent (SC) sera. We observed that most PP sera had antibodies specific to seasonal human CoVs (HCoVs) OC43 and HKU1 spike proteins while the SC sera showed reactivity across all human β-CoVs. Detailed molecular mapping of spike-antibody complexes revealed epitopes that were differentially targeted by preexisting antibodies and SC serum antibodies. Our studies provide an antigenic landscape to β-HCoV spikes in the general population serving as a basis for cross-reactive epitope analyses in SARS-CoV-2-infected individuals.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.