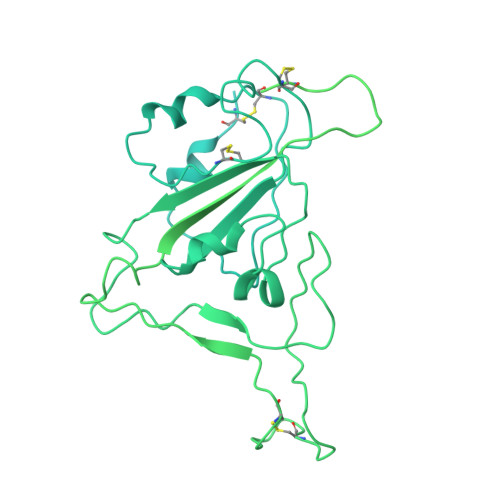
A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Chonira, V., Kwon, Y.D., Gorman, J., Case, J.B., Ku, Z., Simeon, R., Casner, R.G., Harris, D.R., Olia, A.S., Stephens, T., Shapiro, L., Bender, M.F., Boyd, H., Teng, I.T., Tsybovsky, Y., Krammer, F., Zhang, N., Diamond, M.S., Kwong, P.D., An, Z., Chen, Z.(2023) Nat Chem Biol 19: 284-291
- PubMed: 36411391
- DOI: https://doi.org/10.1038/s41589-022-01193-2
- Primary Citation of Related Structures:
7TYZ, 7TZ0, 8DW2, 8DW3 - PubMed Abstract:
We report the engineering and selection of two synthetic proteins-FSR16m and FSR22-for the possible treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. FSR16m and FSR22 are trimeric proteins composed of DARPin SR16m or SR22 fused with a T4 foldon. Despite selection by a spike protein from a now historical SARS-CoV-2 strain, FSR16m and FSR22 exhibit broad-spectrum neutralization of SARS-CoV-2 strains, inhibiting authentic B.1.351, B.1.617.2 and BA.1.1 viruses, with respective IC 50 values of 3.4, 2.2 and 7.4 ng ml -1 for FSR16m. Cryo-EM structures revealed that these DARPins recognize a region of the receptor-binding domain (residues 456, 475, 486, 487 and 489) overlapping a critical portion of the angiotensin-converting enzyme 2 (ACE2)-binding surface. K18-hACE2 transgenic mice inoculated with B.1.617.2 and receiving intranasally administered FSR16m showed less weight loss and 10-100-fold lower viral burden in upper and lower respiratory tracts. The strong and broad neutralization potency makes FSR16m and FSR22 promising candidates for the prevention and treatment of infection by SARS-CoV-2.
Organizational Affiliation:
Department of Microbial Pathogenesis and Immunology, Texas A&M University Health Science Center, Bryan, TX, USA.