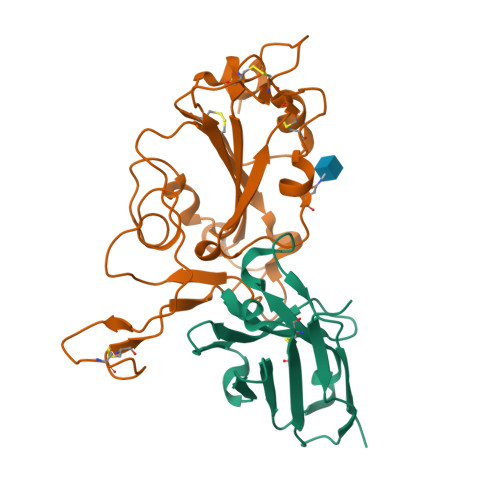
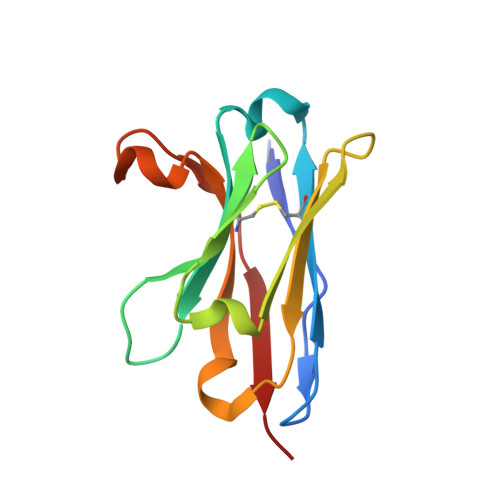
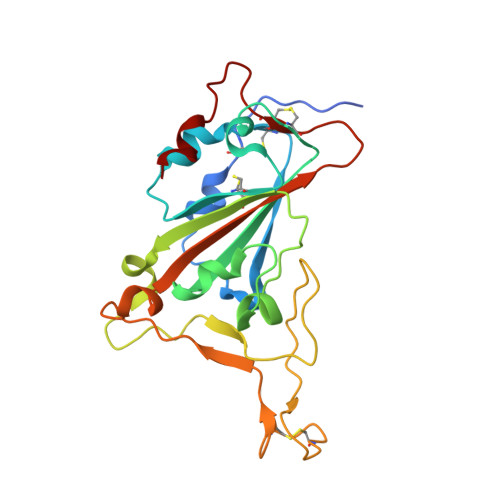
A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Yang, Z., Wang, Y., Jin, Y., Zhu, Y., Wu, Y., Li, C., Kong, Y., Song, W., Tian, X., Zhan, W., Huang, A., Zhou, S., Xia, S., Tian, X., Peng, C., Chen, C., Shi, Y., Hu, G., Du, S., Wang, Y., Xie, Y., Jiang, S., Lu, L., Sun, L., Song, Y., Ying, T.(2021) Signal Transduct Target Ther 6: 378-378
- PubMed: 34732694
- DOI: https://doi.org/10.1038/s41392-021-00810-1
- Primary Citation of Related Structures:
7VNB, 7VNC, 7VND, 7VNE - PubMed Abstract:
The current COVID-19 pandemic has heavily burdened the global public health system and may keep simmering for years. The frequent emergence of immune escape variants have spurred the search for prophylactic vaccines and therapeutic antibodies that confer broad protection against SARS-CoV-2 variants. Here we show that the bivalency of an affinity maturated fully human single-domain antibody (n3113.1-Fc) exhibits exquisite neutralizing potency against SARS-CoV-2 pseudovirus, and confers effective prophylactic and therapeutic protection against authentic SARS-CoV-2 in the host cell receptor angiotensin-converting enzyme 2 (ACE2) humanized mice. The crystal structure of n3113 in complex with the receptor-binding domain (RBD) of SARS-CoV-2, combined with the cryo-EM structures of n3113 and spike ecto-domain, reveals that n3113 binds to the side surface of up-state RBD with no competition with ACE2. The binding of n3113 to this novel epitope stabilizes spike in up-state conformations but inhibits SARS-CoV-2 S mediated membrane fusion, expanding our recognition of neutralization by antibodies against SARS-CoV-2. Binding assay and pseudovirus neutralization assay show no evasion of recently prevalent SARS-CoV-2 lineages, including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) for n3113.1-Fc with Y58L mutation, demonstrating the potential of n3113.1-Fc (Y58L) as a promising candidate for clinical development to treat COVID-19.
Organizational Affiliation:
Department of Pulmonary Medicine, Zhongshan Hospital, Fudan University, Shanghai, 200032, China. yang_zhenlin@fudan.edu.cn.