A dimeric proteomimetic prevents SARS-CoV-2 infection by dimerizing the spike protein.
Khatri, B., Pramanick, I., Malladi, S.K., Rajmani, R.S., Kumar, S., Ghosh, P., Sengupta, N., Rahisuddin, R., Kumar, N., Kumaran, S., Ringe, R.P., Varadarajan, R., Dutta, S., Chatterjee, J.(2022) Nat Chem Biol 18: 1046-1055
- PubMed: 35654847
- DOI: https://doi.org/10.1038/s41589-022-01060-0
- Primary Citation of Related Structures:
7X7N - PubMed Abstract:
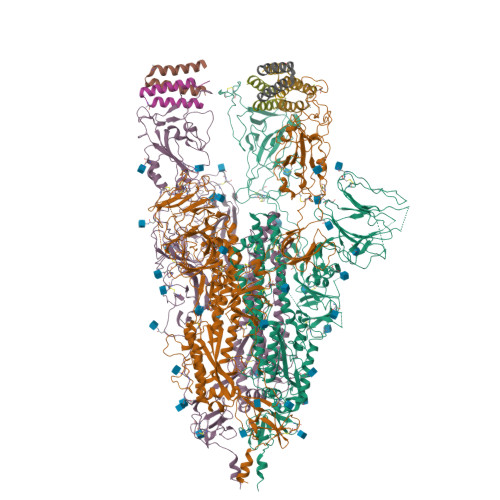
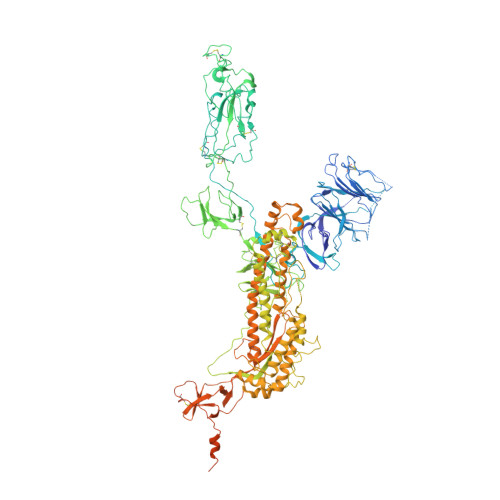
Protein tertiary structure mimetics are valuable tools to target large protein-protein interaction interfaces. Here, we demonstrate a strategy for designing dimeric helix-hairpin motifs from a previously reported three-helix-bundle miniprotein that targets the receptor-binding domain (RBD) of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2). Through truncation of the third helix and optimization of the interhelical loop residues of the miniprotein, we developed a thermostable dimeric helix-hairpin. The dimeric four-helix bundle competes with the human angiotensin-converting enzyme 2 (ACE2) in binding to RBD with 2:2 stoichiometry. Cryogenic-electron microscopy revealed the formation of dimeric spike ectodomain trimer by the four-helix bundle, where all the three RBDs from either spike protein are attached head-to-head in an open conformation, revealing a novel mechanism for virus neutralization. The proteomimetic protects hamsters from high dose viral challenge with replicative SARS-CoV-2 viruses, demonstrating the promise of this class of peptides that inhibit protein-protein interaction through target dimerization.
Organizational Affiliation:
Molecular Biophysics Unit (MBU), Indian Institute of Science, Bangalore, India.