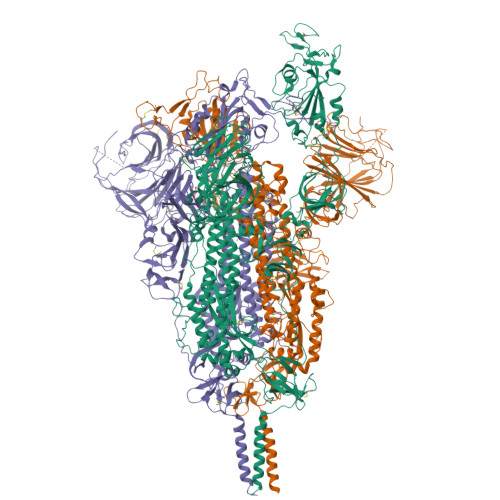
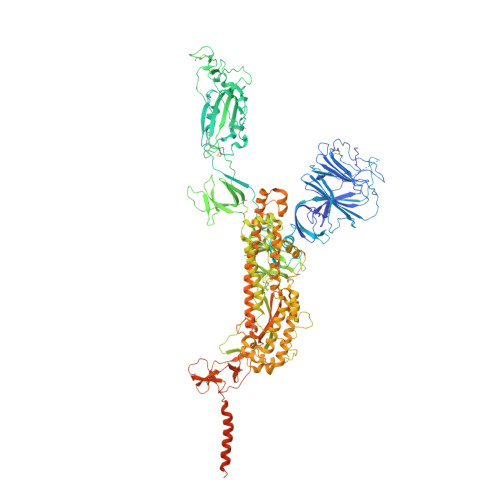
Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Xing, X., Wang, L., Cui, Z., Fu, W., Zheng, T., Qin, L., Ge, P., Qian, A., Wang, N., Yuan, S.(2022) Virol Sin 37: 938-941
- PubMed: 36368512
- DOI: https://doi.org/10.1016/j.virs.2022.11.003
- Primary Citation of Related Structures:
7YBH, 7YBI, 7YBJ, 7YBK, 7YBL, 7YBM, 7YBN - PubMed Abstract:
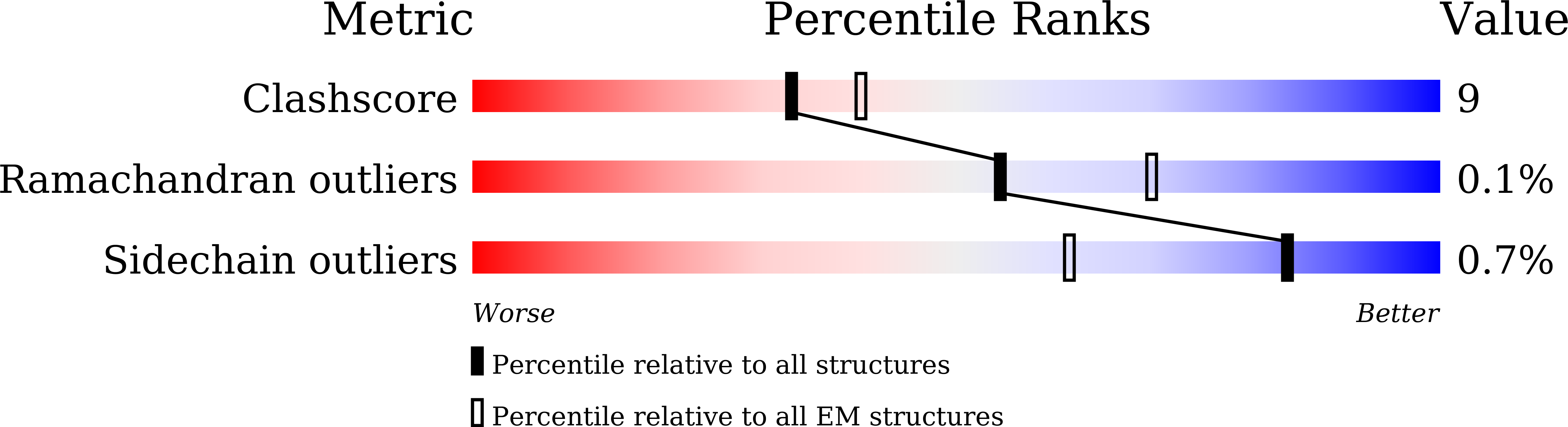
• Deletion of residues 156–157 warps the neighboring beta-sheet and leads NTD and RBD to shift. • T859N stabilizes the packing of the 630 loop motif to make RBD standing transition more difficult. • The overall structures of the closed state S complex from different variants resemble each other. • Mutations in FPPR may affect the overall structure of the trimeric spike protein.
Organizational Affiliation:
College of Veterinary Medicine, Jilin Agricultural University, Changchun, 130118, China; College of Animal Science and Technology, Jilin Provincial Engineering Research Center of Animal Probiotics, Key Lab of Animal Production, Product Quality and Security, Joint Laboratory of Modern Agricultural Technology International Cooperation, Ministry of Education, Jilin Agricultural University, Changchun, 130118, China; CAS Key Laboratory of Infection and Immunity, National Laboratory of Macromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China.