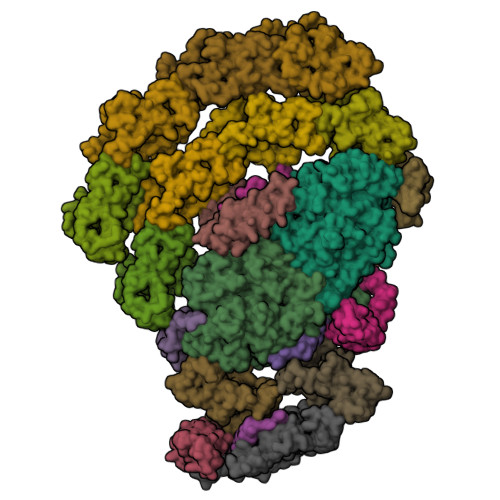
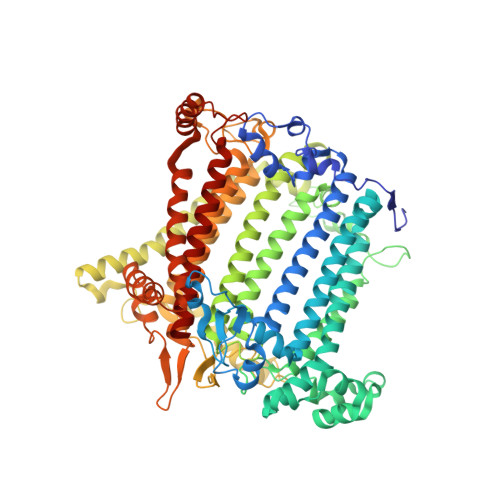
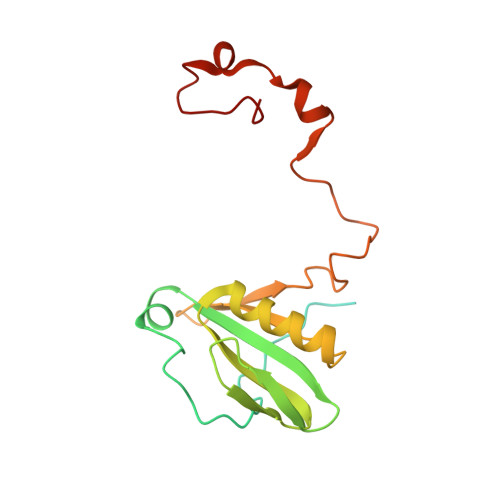
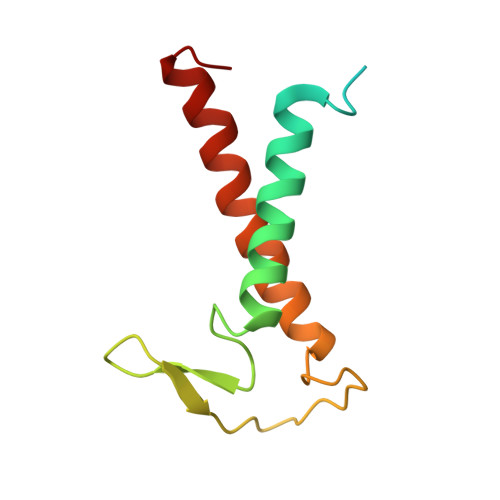
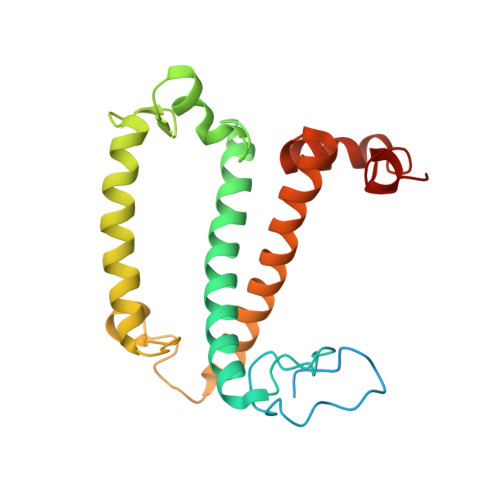
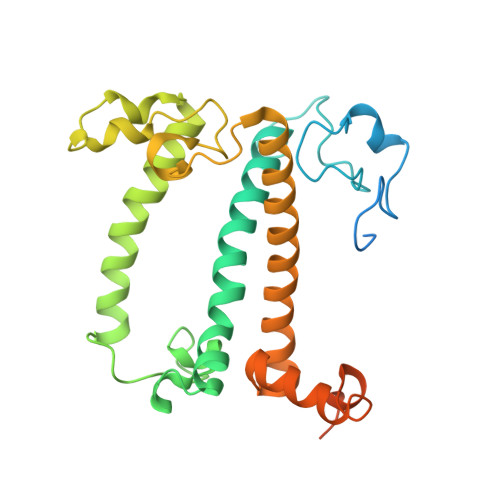
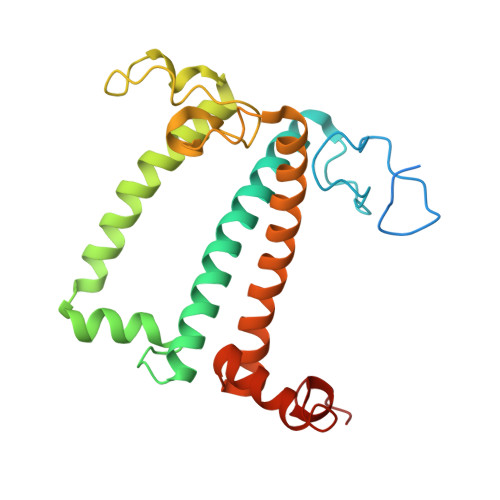
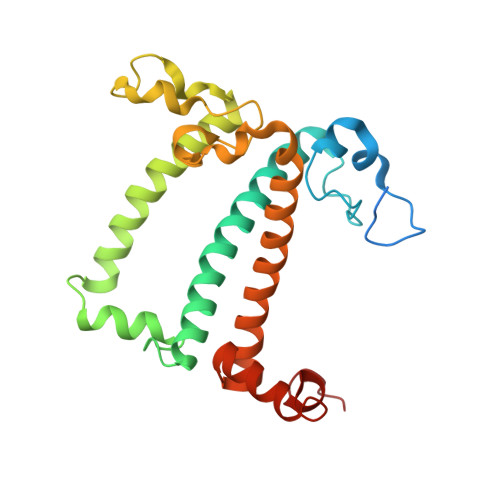
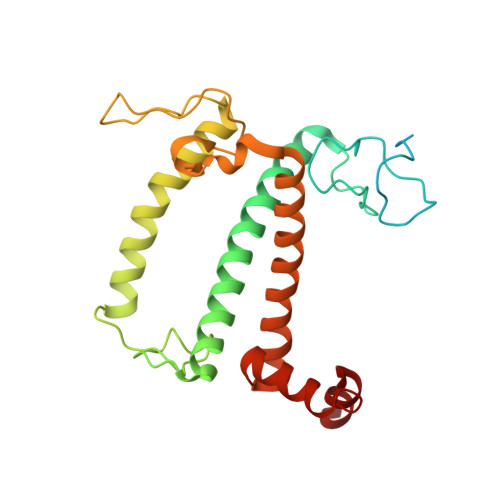
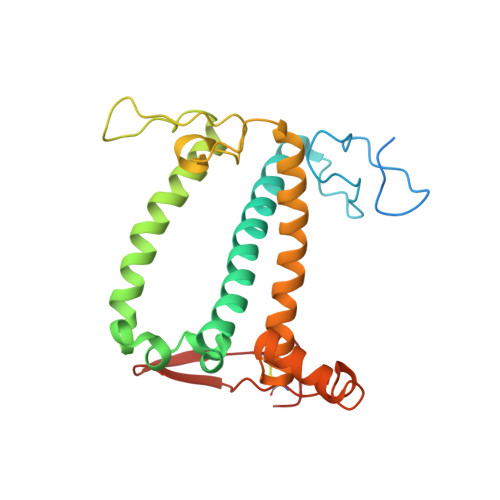
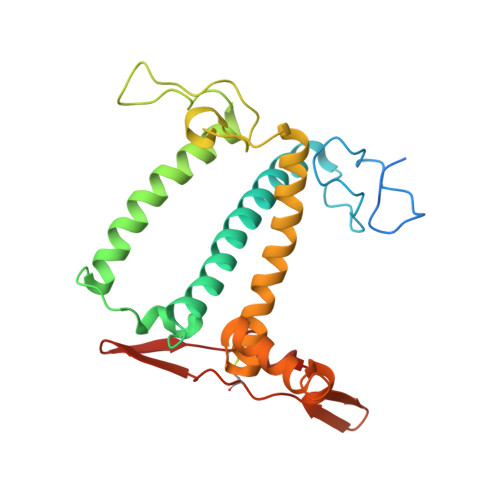
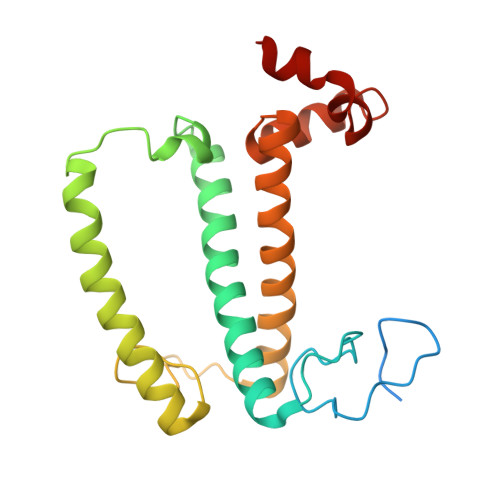
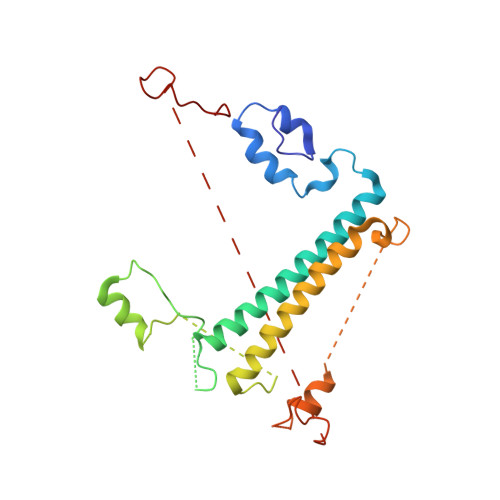
Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Naschberger, A., Mosebach, L., Tobiasson, V., Kuhlgert, S., Scholz, M., Perez-Boerema, A., Ho, T.T.H., Vidal-Meireles, A., Takahashi, Y., Hippler, M., Amunts, A.(2022) Nat Plants 8: 1191-1201
- PubMed: 36229605
- DOI: https://doi.org/10.1038/s41477-022-01253-4
- Primary Citation of Related Structures:
7ZQ9, 7ZQC, 7ZQD, 7ZQE - PubMed Abstract:
Photosystem I (PSI) enables photo-electron transfer and regulates photosynthesis in the bioenergetic membranes of cyanobacteria and chloroplasts. Being a multi-subunit complex, its macromolecular organization affects the dynamics of photosynthetic membranes. Here we reveal a chloroplast PSI from the green alga Chlamydomonas reinhardtii that is organized as a homodimer, comprising 40 protein subunits with 118 transmembrane helices that provide scaffold for 568 pigments. Cryogenic electron microscopy identified that the absence of PsaH and Lhca2 gives rise to a head-to-head relative orientation of the PSI-light-harvesting complex I monomers in a way that is essentially different from the oligomer formation in cyanobacteria. The light-harvesting protein Lhca9 is the key element for mediating this dimerization. The interface between the monomers is lacking PsaH and thus partially overlaps with the surface area that would bind one of the light-harvesting complex II complexes in state transitions. We also define the most accurate available PSI-light-harvesting complex I model at 2.3 Å resolution, including a flexibly bound electron donor plastocyanin, and assign correct identities and orientations to all the pigments, as well as 621 water molecules that affect energy transfer pathways.
Organizational Affiliation:
Science for Life Laboratory, Department of Biochemistry and Biophysics, Stockholm University, Solna, Sweden.