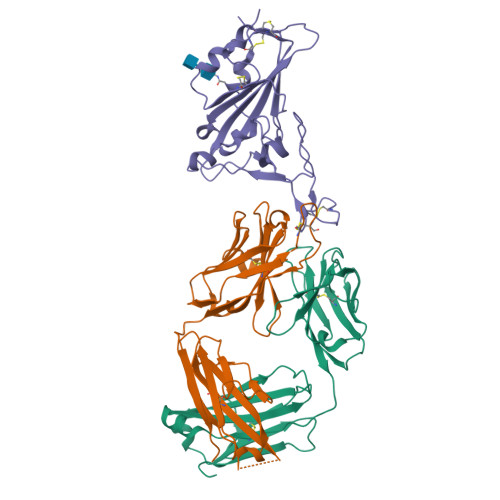
Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Korenkov, M., Zehner, M., Cohen-Dvashi, H., Borenstein-Katz, A., Kottege, L., Janicki, H., Vanshylla, K., Weber, T., Gruell, H., Koch, M., Diskin, R., Kreer, C., Klein, F.(2023) Immunity 56: 2803-2815.e6
- PubMed: 38035879
- DOI: https://doi.org/10.1016/j.immuni.2023.11.004
- Primary Citation of Related Structures:
7B0B - PubMed Abstract:
Somatic hypermutation (SHM) drives affinity maturation and continues over months in SARS-CoV-2-neutralizing antibodies (nAbs). However, several potent SARS-CoV-2 antibodies carry no or only a few mutations, leaving the question of how ongoing SHM affects neutralization unclear. Here, we reverted variable region mutations of 92 antibodies and tested their impact on SARS-CoV-2 binding and neutralization. Reverting higher numbers of mutations correlated with decreasing antibody functionality. However, for some antibodies, including antibodies of the public clonotype VH1-58, neutralization of Wu01 remained unaffected. Although mutations were dispensable for Wu01-induced VH1-58 antibodies to neutralize Alpha, Beta, and Delta variants, they were critical for Omicron BA.1/BA.2 neutralization. We exploited this knowledge to convert the clinical antibody tixagevimab into a BA.1/BA.2 neutralizer. These findings broaden our understanding of SHM as a mechanism that not only improves antibody responses during affinity maturation but also contributes to antibody diversification, thus increasing the chances of neutralizing viral escape variants.
Organizational Affiliation:
Laboratory of Experimental Immunology, Institute of Virology, Faculty of Medicine and University Hospital Cologne, University of Cologne, 50931 Cologne, Germany.