Cross-Reactive SARS-CoV-2 Neutralizing Antibodies From Deep Mining of Early Patient Responses.
Bullen, G., Galson, J.D., Hall, G., Villar, P., Moreels, L., Ledsgaard, L., Mattiuzzo, G., Bentley, E.M., Masters, E.W., Tang, D., Millett, S., Tongue, D., Brown, R., Diamantopoulos, I., Parthiban, K., Tebbutt, C., Leah, R., Chaitanya, K., Ergueta-Carballo, S., Pazeraitis, D., Surade, S.B., Ashiru, O., Crippa, L., Cowan, R., Bowler, M.W., Campbell, J.I., Lee, W.J., Carr, M.D., Matthews, D., Pfeffer, P., Hufton, S.E., Sawmynaden, K., Osbourn, J., McCafferty, J., Karatt-Vellatt, A.(2021) Front Immunol 12: 678570-678570
- PubMed: 34211469
- DOI: https://doi.org/10.3389/fimmu.2021.678570
- Primary Citation of Related Structures:
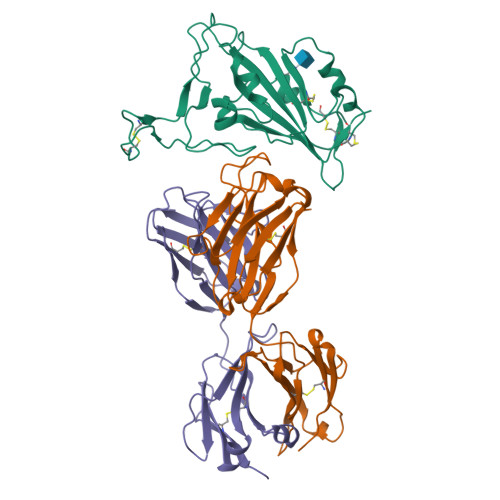
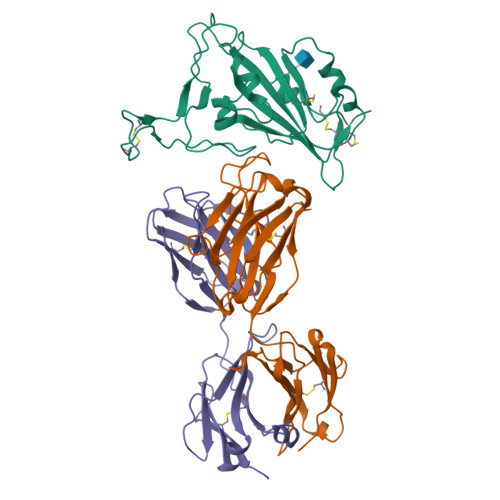
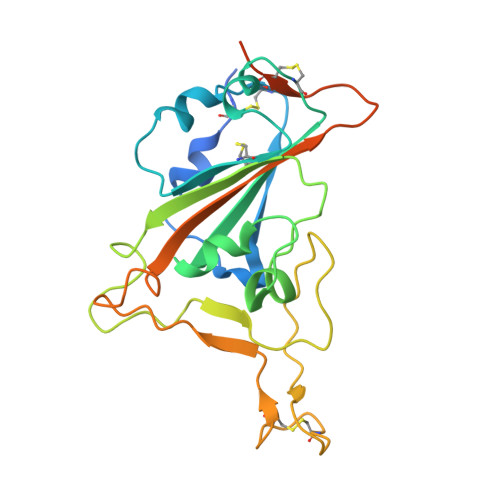
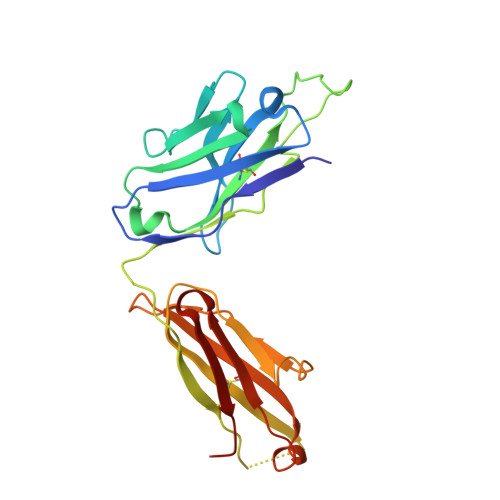
7BNV, 7NP1 - PubMed Abstract:
Passive immunization using monoclonal antibodies will play a vital role in the fight against COVID-19. The recent emergence of viral variants with reduced sensitivity to some current antibodies and vaccines highlights the importance of broad cross-reactivity. This study describes deep-mining of the antibody repertoires of hospitalized COVID-19 patients using phage display technology and B cell receptor (BCR) repertoire sequencing to isolate neutralizing antibodies and gain insights into the early antibody response. This comprehensive discovery approach has yielded a panel of potent neutralizing antibodies which bind distinct viral epitopes including epitopes conserved in SARS-CoV-1. Structural determination of a non-ACE2 receptor blocking antibody reveals a previously undescribed binding epitope, which is unlikely to be affected by the mutations in any of the recently reported major viral variants including B.1.1.7 (from the UK), B.1.351 (from South Africa) and B.1.1.28 (from Brazil). Finally, by combining sequences of the RBD binding and neutralizing antibodies with the B cell receptor repertoire sequencing, we also describe a highly convergent early antibody response. Similar IgM-derived sequences occur within this study group and also within patient responses described by multiple independent studies published previously.
Organizational Affiliation:
IONTAS Ltd., Cambridge, United Kingdom.