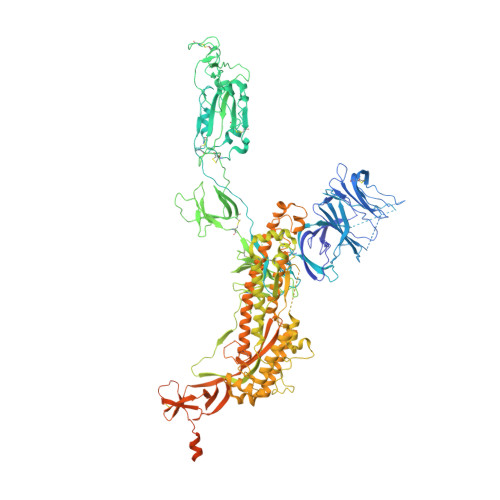
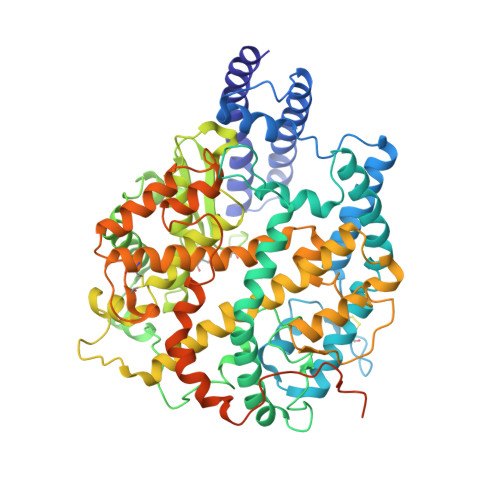
Engineered trimeric ACE2 binds viral spike protein and locks it in "Three-up" conformation to potently inhibit SARS-CoV-2 infection.
Guo, L., Bi, W., Wang, X., Xu, W., Yan, R., Zhang, Y., Zhao, K., Li, Y., Zhang, M., Cai, X., Jiang, S., Xie, Y., Zhou, Q., Lu, L., Dang, B.(2021) Cell Res 31: 98-100
- PubMed: 33177651
- DOI: https://doi.org/10.1038/s41422-020-00438-w
- Primary Citation of Related Structures:
7CT5
Organizational Affiliation:
Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, Zhejiang, 310024, China.