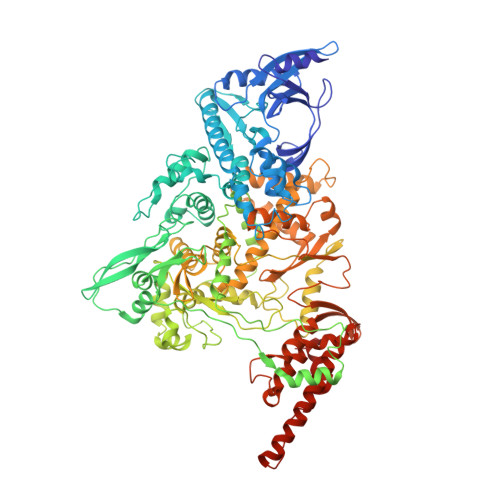
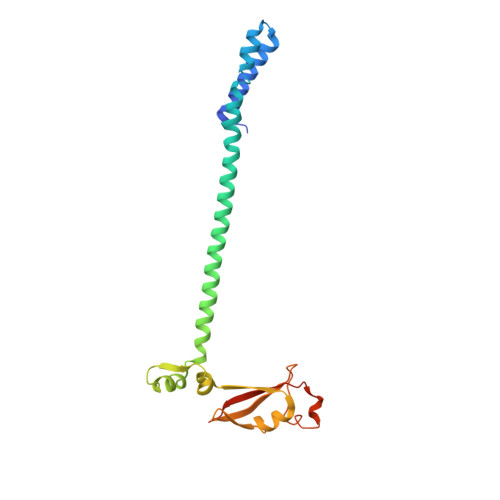
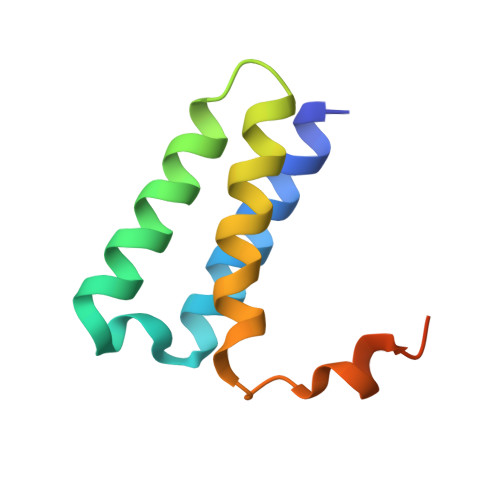
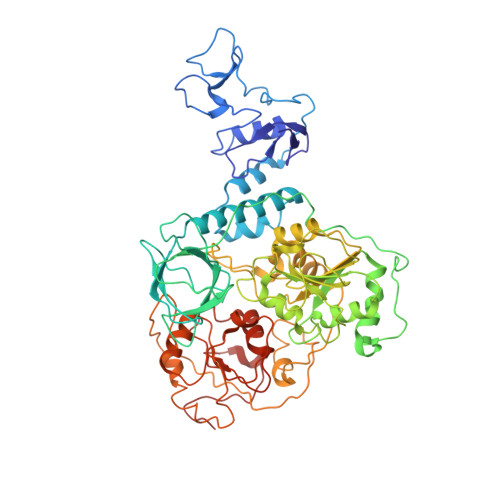
Architecture of a SARS-CoV-2 mini replication and transcription complex.
Yan, L., Zhang, Y., Ge, J., Zheng, L., Gao, Y., Wang, T., Jia, Z., Wang, H., Huang, Y., Li, M., Wang, Q., Rao, Z., Lou, Z.(2020) Nat Commun 11: 5874-5874
- PubMed: 33208736
- DOI: https://doi.org/10.1038/s41467-020-19770-1
- Primary Citation of Related Structures:
7CXM, 7CXN - PubMed Abstract:
Non-structural proteins (nsp) constitute the SARS-CoV-2 replication and transcription complex (RTC) to play a pivotal role in the virus life cycle. Here we determine the atomic structure of a SARS-CoV-2 mini RTC, assembled by viral RNA-dependent RNA polymerase (RdRp, nsp12) with a template-primer RNA, nsp7 and nsp8, and two helicase molecules (nsp13-1 and nsp13-2), by cryo-electron microscopy. Two groups of mini RTCs with different conformations of nsp13-1 are identified. In both of them, nsp13-1 stabilizes overall architecture of the mini RTC by contacting with nsp13-2, which anchors the 5'-extension of RNA template, as well as interacting with nsp7-nsp8-nsp12-RNA. Orientation shifts of nsp13-1 results in its variable interactions with other components in two forms of mini RTC. The mutations on nsp13-1:nsp12 and nsp13-1:nsp13-2 interfaces prohibit the enhancement of helicase activity achieved by mini RTCs. These results provide an insight into how helicase couples with polymerase to facilitate its function in virus replication and transcription.
Organizational Affiliation:
MOE Key Laboratory of Protein Science, School of Medicine, Tsinghua University, Beijing, China.