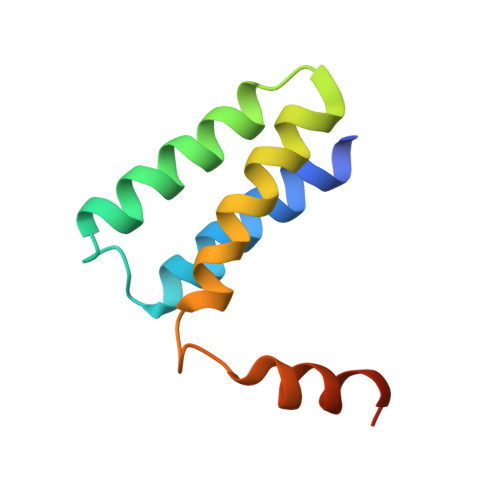
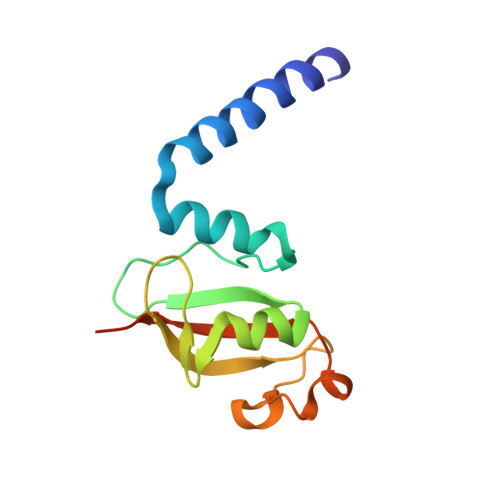
Nonstructural protein 7 and 8 complexes of SARS-CoV-2.
Zhang, C., Li, L., He, J., Chen, C., Su, D.(2021) Protein Sci 30: 873-881
- PubMed: 33594727
- DOI: https://doi.org/10.1002/pro.4046
- Primary Citation of Related Structures:
7DCD - PubMed Abstract:
The pandemic outbreak of coronavirus disease 2019 (COVID-19) across the world has led to millions of infection cases and caused a global public health crisis. Current research suggests that SARS-CoV-2 is a highly contagious coronavirus that spreads rapidly through communities. To understand the mechanisms of viral replication, it is imperative to investigate coronavirus viral replicase, a huge protein complex comprising up to 16 viral nonstructural and associated host proteins, which is the most promising antiviral target for inhibiting viral genome replication and transcription. Recently, several components of the viral replicase complex in SARS-CoV-2 have been solved to provide a basis for the design of new antiviral therapeutics. Here, we report the crystal structure of the SARS-CoV-2 nsp7+8 tetramer, which comprises two copies of each protein representing nsp7's full-length and the C-terminus of nsp8 owing to N-terminus proteolysis during the process of crystallization. We also identified a long helical extension and highly flexible N-terminal domain of nsp8, which is preferred for interacting with single-stranded nucleic acids.
Organizational Affiliation:
State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, and Collaborative Innovation Center for Biotherapy, Chengdu, China.