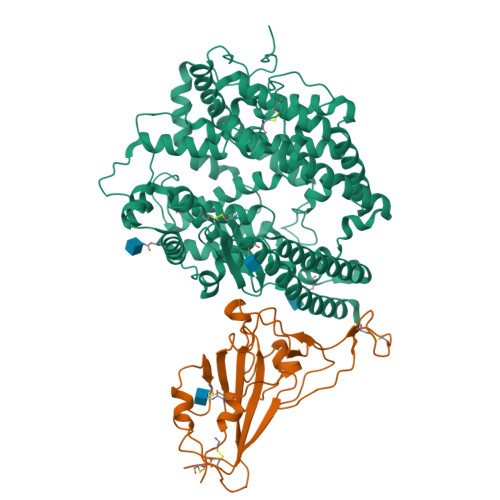
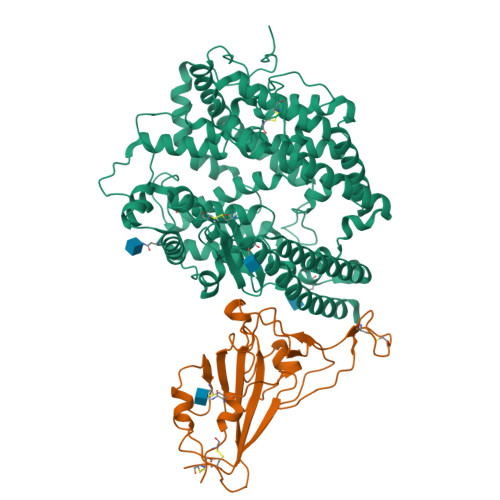
Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Niu, S., Wang, J., Bai, B., Wu, L., Zheng, A., Chen, Q., Du, P., Han, P., Zhang, Y., Jia, Y., Qiao, C., Qi, J., Tian, W.X., Wang, H.W., Wang, Q., Gao, G.F.(2021) EMBO J 40: e107786-e107786
- PubMed: 34018203
- DOI: https://doi.org/10.15252/embj.2021107786
- Primary Citation of Related Structures:
7DDO, 7DDP - PubMed Abstract:
Pangolins have been suggested as potential reservoir of zoonotic viruses, including SARS-CoV-2 causing the global COVID-19 outbreak. Here, we study the binding of two SARS-CoV-2-like viruses isolated from pangolins, GX/P2V/2017 and GD/1/2019, to human angiotensin-converting enzyme 2 (hACE2), the receptor of SARS-CoV-2. We find that the spike protein receptor-binding domain (RBD) of pangolin CoVs binds to hACE2 as efficiently as the SARS-CoV-2 RBD in vitro. Furthermore, incorporation of pangolin CoV RBDs allows entry of pseudotyped VSV particles into hACE2-expressing cells. A screen for binding of pangolin CoV RBDs to ACE2 orthologs from various species suggests a broader host range than that of SARS-CoV-2. Additionally, cryo-EM structures of GX/P2V/2017 and GD/1/2019 RBDs in complex with hACE2 show their molecular binding in modes similar to SARS-CoV-2 RBD. Introducing the Q498H substitution found in pangolin CoVs into the SARS-CoV-2 RBD expands its binding capacity to ACE2 homologs of mouse, rat, and European hedgehog. These findings suggest that these two pangolin CoVs may infect humans, highlighting the necessity of further surveillance of pangolin CoVs.
Organizational Affiliation:
College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, China.