The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Wang, H., Pei, R., Li, X., Deng, W., Xing, S., Zhang, Y., Zhang, C., He, S., Sun, H., Xiao, S., Xiong, J., Zhang, Y., Chen, X., Wang, Y., Guo, Y., Zhang, B., Shang, L.(2022) Eur J Med Chem 238: 114458-114458
- PubMed: 35635946
- DOI: https://doi.org/10.1016/j.ejmech.2022.114458
- Primary Citation of Related Structures:
7DGB, 7DGF, 7DGG, 7DGH, 7DGI, 7DHJ - PubMed Abstract:
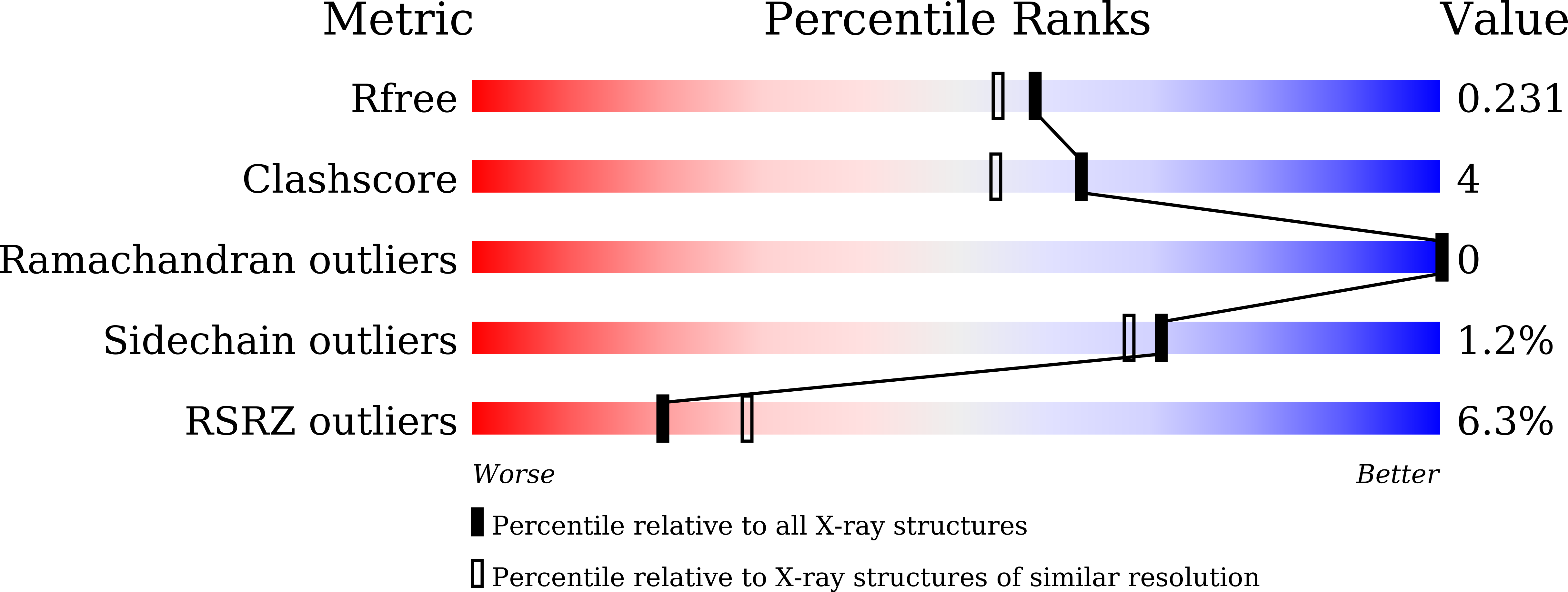
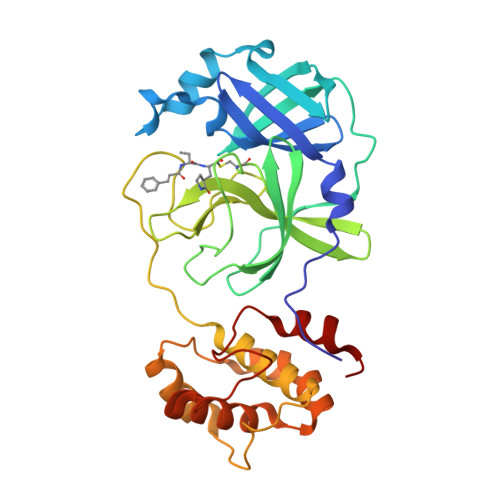
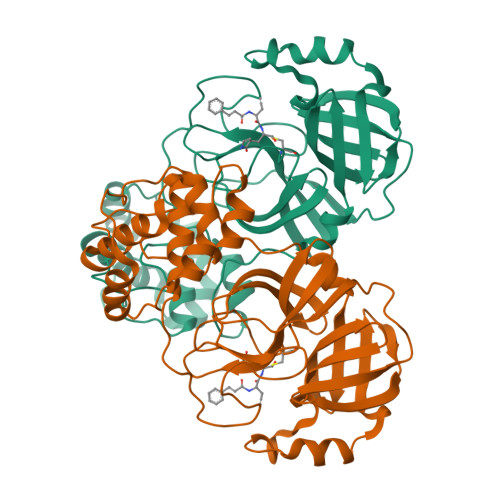
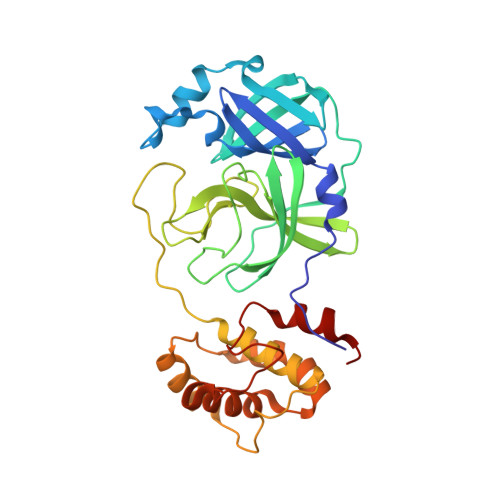
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), as the pathogen of coronavirus disease 2019 (COVID-19), has infected millions of people and took hundreds of thousands of lives. Unfortunately, there is deficiency of effective medicines to prevent or treat COVID-19. 3C like protease (3CL Pro ) of SARS-CoV-2 is essential to the viral replication and transcription, and is an attractive target to develop anti-SARS-CoV-2 agents. Targeting on the 3CL Pro , we screened our protease inhibitor library and obtained compound 10a as hit to weakly inhibit the SARS-CoV-2 3CL Pro , and determined the co-crystal structure of 10a and the protease. Based on the deep understanding on the protein-ligand complexes between the hit and SARS-CoV-2 3CL Pro , we designed a series of peptidomimetic inhibitors, with outstanding inhibitory activity against SARS-CoV-2 3CL Pro and excellent anti-viral potency against SARS-CoV-2. The protein-ligand complexes of the other key inhibitors with SARS-CoV-2 3CL Pro were explicitly described by the X-ray co-crystal study. All such results suggest these peptidomimetic inhibitors could be further applied as encouraging drug candidates.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy, KLMDASR of Tianjin and Drug Discovery Center for Infectious Disease, Nankai University, Tianjin, 300353, People's Republic of China.