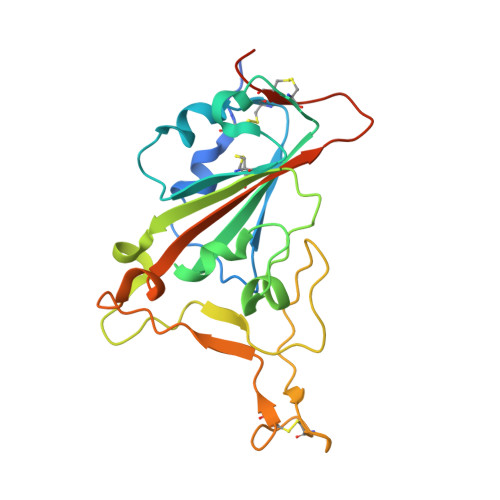
Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.
Jiang, W., Wang, J., Jiao, S., Gu, C., Xu, W., Chen, B., Wang, R., Chen, H., Xie, Y., Wang, A., Li, G., Zeng, D., Zhang, J., Zhang, M., Wang, S., Wang, M., Gui, X.(2021) MAbs 13: 1953683-1953683
- PubMed: 34313527
- DOI: https://doi.org/10.1080/19420862.2021.1953683
- Primary Citation of Related Structures:
7DPM - PubMed Abstract:
The global pandemic of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in widespread social and economic disruption. Effective interventions are urgently needed for the prevention and treatment of COVID-19. Neutralizing monoclonal antibodies (mAbs) have demonstrated their prophylactic and therapeutic efficacy against SARS-CoV-2, and several have been granted authorization for emergency use. Here, we discover and characterize a fully human cross-reactive mAb, MW06, which binds to both SARS-CoV-2 and SARS-CoV spike receptor-binding domain (RBD) and disrupts their interaction with angiotensin-converting enzyme 2 (ACE2) receptors. Potential neutralization activity of MW06 was observed against both SARS-CoV-2 and SARS-CoV in different assays. The complex structure determination and epitope alignment of SARS-CoV-2 RBD/MW06 revealed that the epitope recognized by MW06 is highly conserved among SARS-related coronavirus strains, indicating the potential broad neutralization activity of MW06. In in vitro assays, no antibody-dependent enhancement (ADE) of SARS-CoV-2 infection was observed for MW06. In addition, MW06 recognizes a different epitope from MW05, which shows high neutralization activity and has been in a Phase 2 clinical trial, supporting the development of the cocktail of MW05 and MW06 to prevent against future escaping variants. MW06 alone and the cocktail show good effects in preventing escape mutations, including a series of variants of concern, B.1.1.7, P.1, B.1.351, and B.1.617.1. These findings suggest that MW06 recognizes a conserved epitope on SARS-CoV-2, which provides insights for the development of a universal antibody-based therapy against SARS-related coronavirus and emerging variant strains, and may be an effective anti-SARS-CoV-2 agent.
Organizational Affiliation:
Department of Antibody Discovery and Development, Mabwell (Shanghai) Bioscience Co., Ltd, Shanghai, China.