A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Onodera, T., Kita, S., Adachi, Y., Moriyama, S., Sato, A., Nomura, T., Sakakibara, S., Inoue, T., Tadokoro, T., Anraku, Y., Yumoto, K., Tian, C., Fukuhara, H., Sasaki, M., Orba, Y., Shiwa, N., Iwata, N., Nagata, N., Suzuki, T., Sasaki, J., Sekizuka, T., Tonouchi, K., Sun, L., Fukushi, S., Satofuka, H., Kazuki, Y., Oshimura, M., Kurosaki, T., Kuroda, M., Matsuura, Y., Suzuki, T., Sawa, H., Hashiguchi, T., Maenaka, K., Takahashi, Y.(2021) Immunity 54: 2385
- PubMed: 34508662
- DOI: https://doi.org/10.1016/j.immuni.2021.08.025
- Primary Citation of Related Structures:
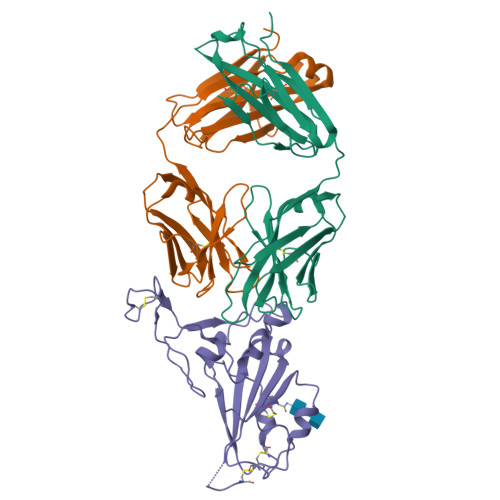
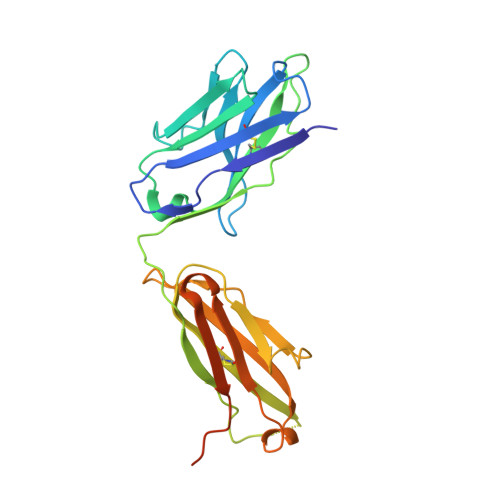
7E5O - PubMed Abstract:
Potent neutralizing SARS-CoV-2 antibodies often target the spike protein receptor-binding site (RBS), but the variability of RBS epitopes hampers broad neutralization of multiple sarbecoviruses and drifted viruses. Here, using humanized mice, we identified an RBS antibody with a germline V H gene that potently neutralized SARS-related coronaviruses, including SARS-CoV and SARS-CoV-2 variants. X-ray crystallography revealed coordinated recognition by the heavy chain of non-RBS conserved sites and the light chain of RBS with a binding angle mimicking the angiotensin-converting enzyme 2 (ACE2) receptor. The minimum footprints in the hypervariable region of RBS contributed to the breadth of neutralization, which was enhanced by immunoglobulin G3 (IgG3) class switching. The coordinated binding resulted in broad neutralization of SARS-CoV and emerging SARS-CoV-2 variants of concern. Low-dose therapeutic antibody treatment in hamsters reduced the virus titers and morbidity during SARS-CoV-2 challenge. The structural basis for broad neutralizing activity may inform the design of a broad spectrum of therapeutics and vaccines.
Organizational Affiliation:
Research Center for Drug and Vaccine Development, National Institute of Infectious Diseases, Tokyo 162-8640, Japan.