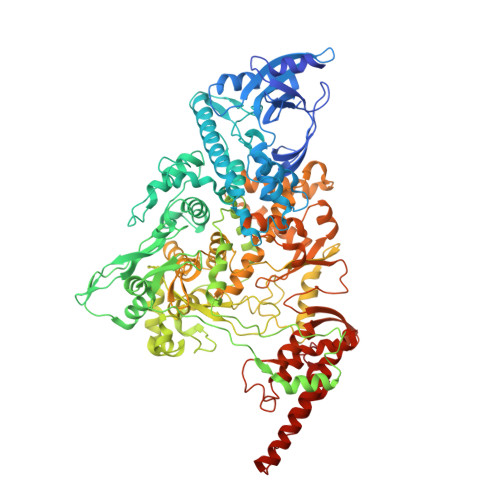
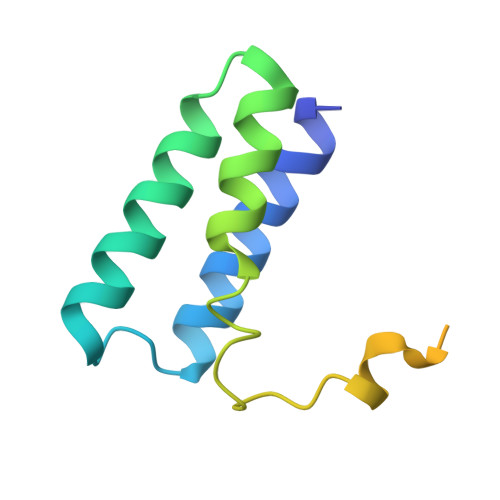
A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Shannon, A., Fattorini, V., Sama, B., Selisko, B., Feracci, M., Falcou, C., Gauffre, P., El Kazzi, P., Delpal, A., Decroly, E., Alvarez, K., Eydoux, C., Guillemot, J.C., Moussa, A., Good, S.S., La Colla, P., Lin, K., Sommadossi, J.P., Zhu, Y., Yan, X., Shi, H., Ferron, F., Canard, B.(2022) Nat Commun 13: 621-621
- PubMed: 35110538
- DOI: https://doi.org/10.1038/s41467-022-28113-1
- Primary Citation of Related Structures:
7ED5 - PubMed Abstract:
The guanosine analog AT-527 represents a promising candidate against Severe Acute Respiratory Syndrome coronavirus type 2 (SARS-CoV-2). AT-527 recently entered phase III clinical trials for the treatment of COVID-19. Once in cells, AT-527 is converted into its triphosphate form, AT-9010, that presumably targets the viral RNA-dependent RNA polymerase (RdRp, nsp12), for incorporation into viral RNA. Here we report a 2.98 Å cryo-EM structure of the SARS-CoV-2 nsp12-nsp7-nsp8 2 -RNA complex, showing AT-9010 bound at three sites of nsp12. In the RdRp active-site, one AT-9010 is incorporated at the 3' end of the RNA product strand. Its modified ribose group (2'-fluoro, 2'-methyl) prevents correct alignment of the incoming NTP, in this case a second AT-9010, causing immediate termination of RNA synthesis. The third AT-9010 is bound to the N-terminal domain of nsp12 - known as the NiRAN. In contrast to native NTPs, AT-9010 is in a flipped orientation in the active-site, with its guanine base unexpectedly occupying a previously unnoticed cavity. AT-9010 outcompetes all native nucleotides for NiRAN binding, inhibiting its nucleotidyltransferase activity. The dual mechanism of action of AT-527 at both RdRp and NiRAN active sites represents a promising research avenue against COVID-19.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, CNRS and Aix Marseille Université, UMR 7257, Polytech Case 925, 13009, Marseille, France.