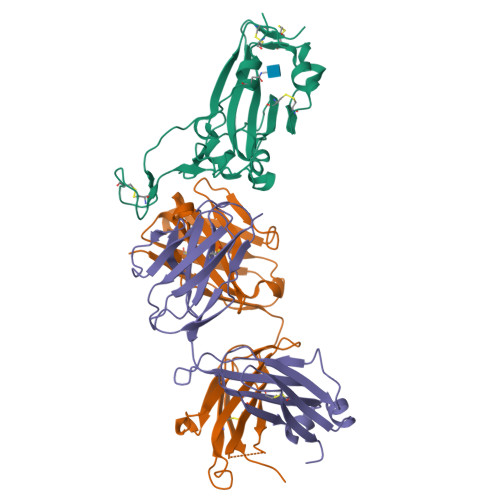
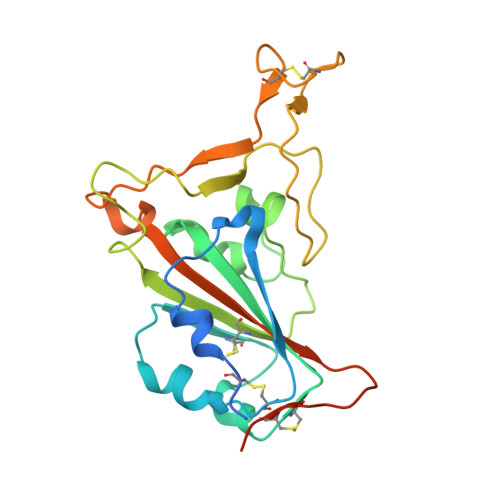
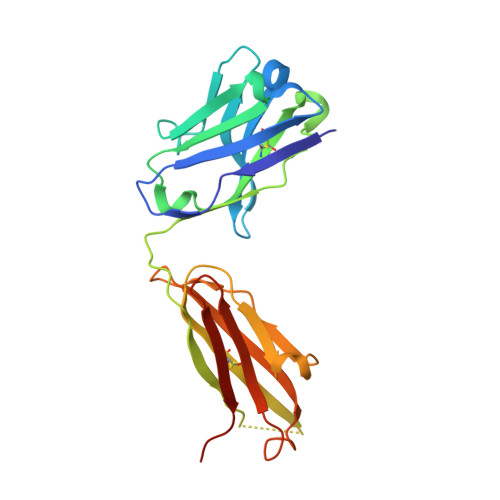
Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Xu, H., Wang, B., Zhao, T.N., Liang, Z.T., Peng, T.B., Song, X.H., Wu, J.J., Wang, Y.C., Su, X.D.(2021) Cell Res 31: 1126-1129
- PubMed: 34480123
- DOI: https://doi.org/10.1038/s41422-021-00554-1
- Primary Citation of Related Structures:
7EJY, 7EJZ, 7EK0, 7F6Y, 7F6Z
Organizational Affiliation:
State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, and Biomedical Pioneering Innovation Center (BIOPIC), Peking University, Beijing, China.