Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Han, P., Su, C., Zhang, Y., Bai, C., Zheng, A., Qiao, C., Wang, Q., Niu, S., Chen, Q., Zhang, Y., Li, W., Liao, H., Li, J., Zhang, Z., Cho, H., Yang, M., Rong, X., Hu, Y., Huang, N., Yan, J., Wang, Q., Zhao, X., Gao, G.F., Qi, J.(2021) Nat Commun 12: 6103-6103
- PubMed: 34671049
- DOI: https://doi.org/10.1038/s41467-021-26401-w
- Primary Citation of Related Structures:
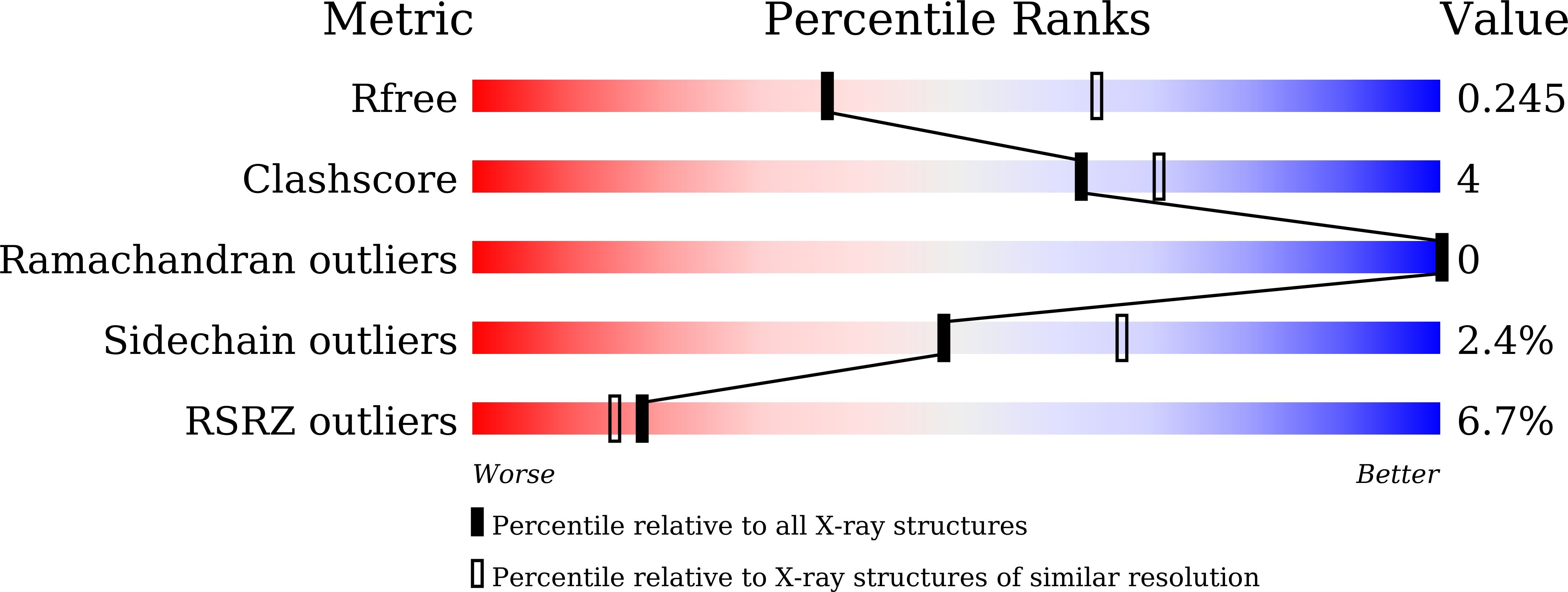
7EKC, 7EKE, 7EKF, 7EKG, 7EKH - PubMed Abstract:
Multiple SARS-CoV-2 variants of concern (VOCs) have been emerging and some have been linked to an increase in case numbers globally. However, there is yet a lack of understanding of the molecular basis for the interactions between the human ACE2 (hACE2) receptor and these VOCs. Here we examined several VOCs including Alpha, Beta, and Gamma, and demonstrate that five variants receptor-binding domain (RBD) increased binding affinity for hACE2, and four variants pseudoviruses increased entry into susceptible cells. Crystal structures of hACE2-RBD complexes help identify the key residues facilitating changes in hACE2 binding affinity. Additionally, soluble hACE2 protein efficiently prevent most of the variants pseudoviruses. Our findings provide important molecular information and may help the development of novel therapeutic and prophylactic agents targeting these emerging mutants.
Organizational Affiliation:
CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China.