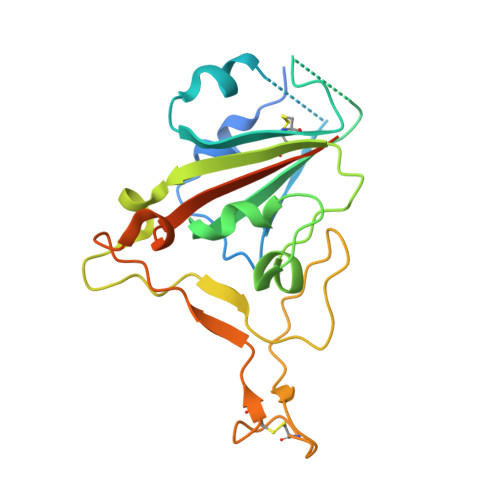
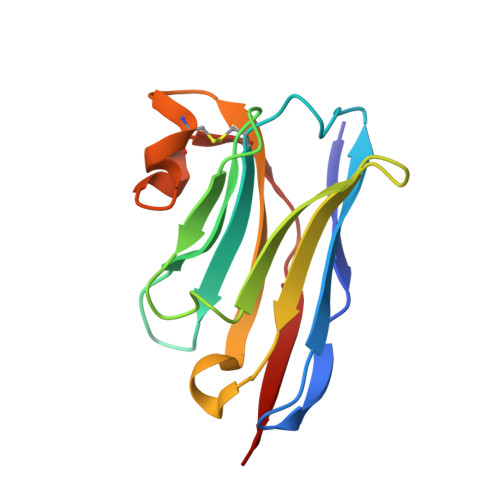
Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Shi, Z., Li, X., Wang, L., Sun, Z., Zhang, H., Chen, X., Cui, Q., Qiao, H., Lan, Z., Zhang, X., Li, X., Li, L., Xu, J., Gong, R., Fan, C., Geng, Y.(2022) Structure 30: 707-720.e5
- PubMed: 35276082
- DOI: https://doi.org/10.1016/j.str.2022.02.011
- Primary Citation of Related Structures:
7FAU - PubMed Abstract:
Because of the evolutionary variants of SARS-CoV-2, development of broad-spectrum neutralizing antibodies resilient to virus escape is urgently needed. We identified a group of high-affinity nanobodies from camels immunized with receptor-binding domain (RBD) of SARS-CoV-2 spike protein and resolved the structures of two non-competing nanobodies (NB1A7 and NB1B11) in complex with RBD using X-ray crystallography. The structures show that NB1A7 targets the highly conserved cryptic epitope shared by SARS-CoV-2 variants and some other coronaviruses and blocks ACE2 receptor attachment of the spike protein, and NB1B11 epitope overlaps with the contacting surface of ACE2 and is different from the binding site of NB1A7. These two nanobodies were covalently linked into multivalent and bi-paratopic formats, which significantly improved the avidity and neutralization potency and may further inhibit viral escape. The results contribute to the structure-guided design of antibodies against future variants of SARS-CoV-2 virus to combat coronavirus epidemics and pandemics.
Organizational Affiliation:
The CAS Key Laboratory of Receptor Research, Stake Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; Department of Biopharmaceutics, College of Food Science and Technology, Shanghai Ocean University, Shanghai 201306, China.