Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Xiang, Y., Nambulli, S., Xiao, Z., Liu, H., Sang, Z., Duprex, W.P., Schneidman-Duhovny, D., Zhang, C., Shi, Y.(2020) Science 370: 1479-1484
- PubMed: 33154108
- DOI: https://doi.org/10.1126/science.abe4747
- Primary Citation of Related Structures:
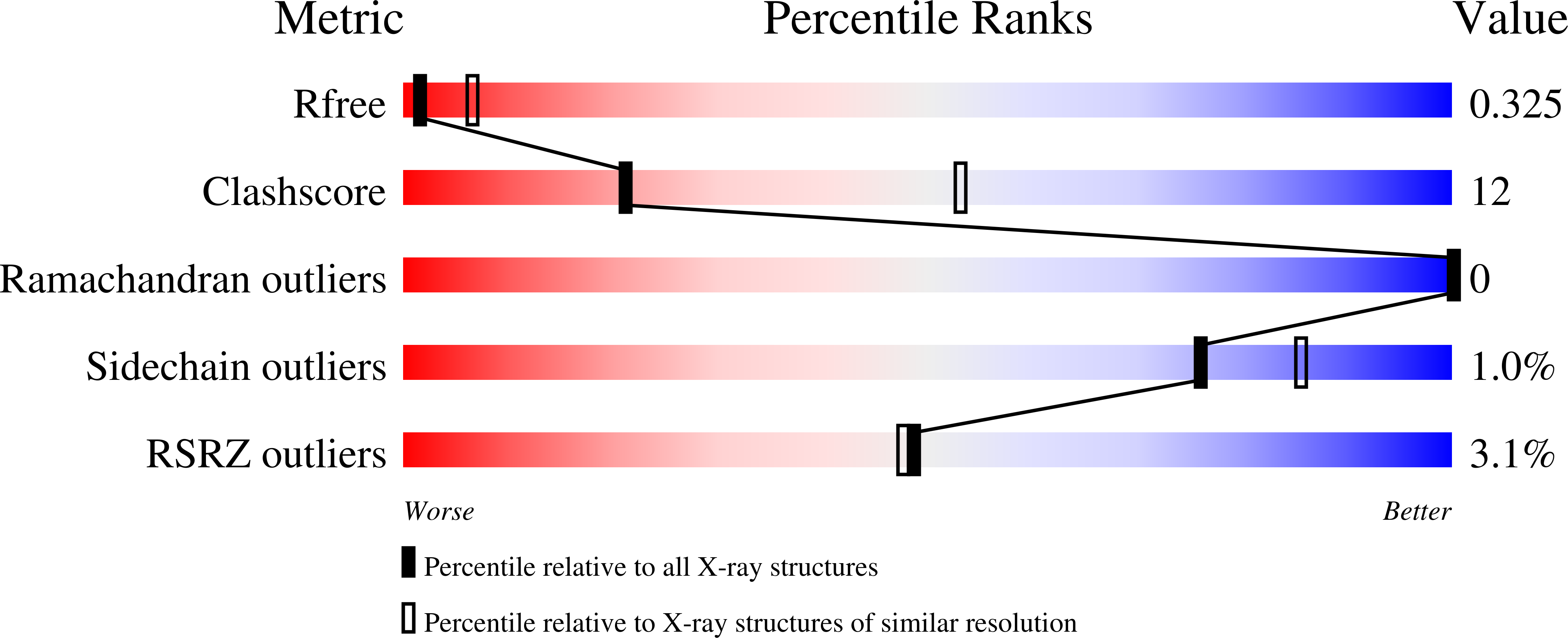
7JVB - PubMed Abstract:
Cost-effective, efficacious therapeutics are urgently needed to combat the COVID-19 pandemic. In this study, we used camelid immunization and proteomics to identify a large repertoire of highly potent neutralizing nanobodies (Nbs) to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein receptor binding domain (RBD). We discovered Nbs with picomolar to femtomolar affinities that inhibit viral infection at concentrations below the nanograms-per-milliliter level, and we determined a structure of one of the most potent Nbs in complex with the RBD. Structural proteomics and integrative modeling revealed multiple distinct and nonoverlapping epitopes and indicated an array of potential neutralization mechanisms. We bioengineered multivalent Nb constructs that achieved ultrahigh neutralization potency (half-maximal inhibitory concentration as low as 0.058 ng/ml) and may prevent mutational escape. These thermostable Nbs can be rapidly produced in bulk from microbes and resist lyophilization and aerosolization.
Organizational Affiliation:
Department of Cell Biology, University of Pittsburgh, Pittsburgh, PA, USA.