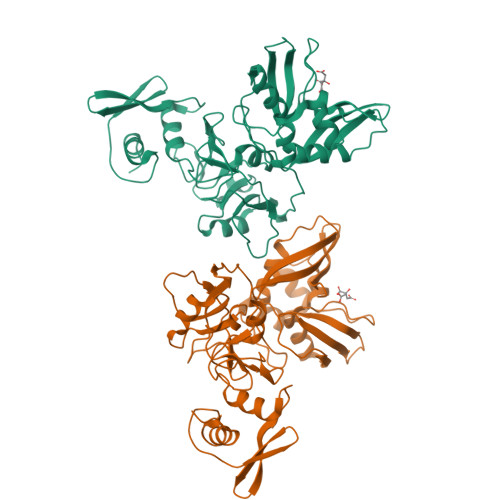
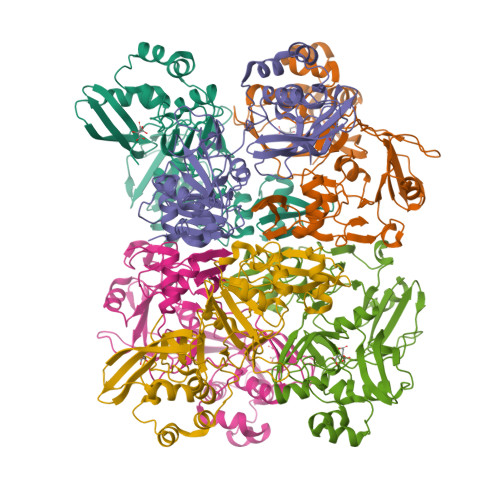
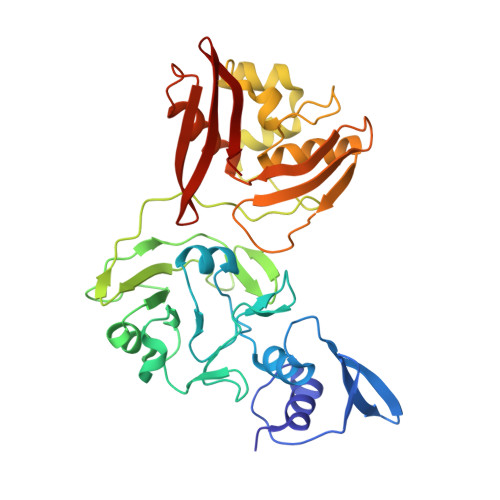
Room-temperature structural studies of SARS-CoV-2 protein NendoU with an X-ray free-electron laser.
Jernigan, R.J., Logeswaran, D., Doppler, D., Nagaratnam, N., Sonker, M., Yang, J.H., Ketawala, G., Martin-Garcia, J.M., Shelby, M.L., Grant, T.D., Mariani, V., Tolstikova, A., Sheikh, M.Z., Yung, M.C., Coleman, M.A., Zaare, S., Kaschner, E.K., Rabbani, M.T., Nazari, R., Zacks, M.A., Hayes, B., Sierra, R.G., Hunter, M.S., Lisova, S., Batyuk, A., Kupitz, C., Boutet, S., Hansen, D.T., Kirian, R.A., Schmidt, M., Fromme, R., Frank, M., Ros, A., Chen, J.J., Botha, S., Fromme, P.(2022) Structure
- PubMed: 36630960
- DOI: https://doi.org/10.1016/j.str.2022.12.009
- Primary Citation of Related Structures:
7K9P - PubMed Abstract:
NendoU from SARS-CoV-2 is responsible for the virus's ability to evade the innate immune system by cleaving the polyuridine leader sequence of antisense viral RNA. Here we report the room-temperature structure of NendoU, solved by serial femtosecond crystallography at an X-ray free-electron laser to 2.6 Å resolution. The room-temperature structure provides insight into the flexibility, dynamics, and other intrinsic properties of NendoU, with indications that the enzyme functions as an allosteric switch. Functional studies examining cleavage specificity in solution and in crystals support the uridine-purine cleavage preference, and we demonstrate that enzyme activity is fully maintained in crystal form. Optimizing the purification of NendoU and identifying suitable crystallization conditions set the benchmark for future time-resolved serial femtosecond crystallography studies. This could advance the design of antivirals with higher efficacy in treating coronaviral infections, since drugs that block allosteric conformational changes are less prone to drug resistance.
Organizational Affiliation:
Biodesign Center for Applied Structural Discovery, Arizona State University, Tempe, AZ 85287-5001, USA; School of Molecular Sciences, Arizona State University, Tempe, AZ 85287-1604, USA.