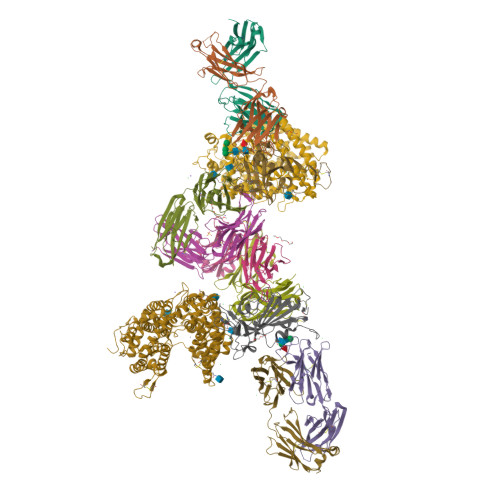
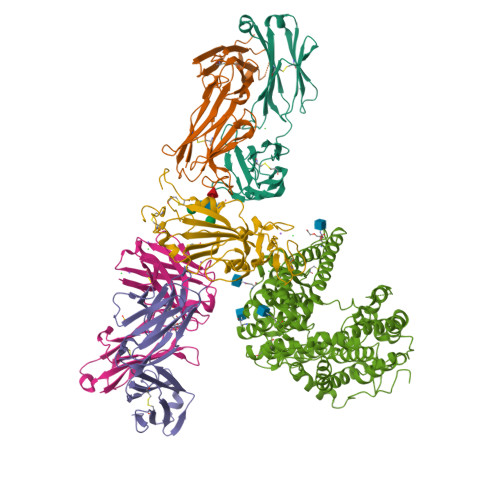
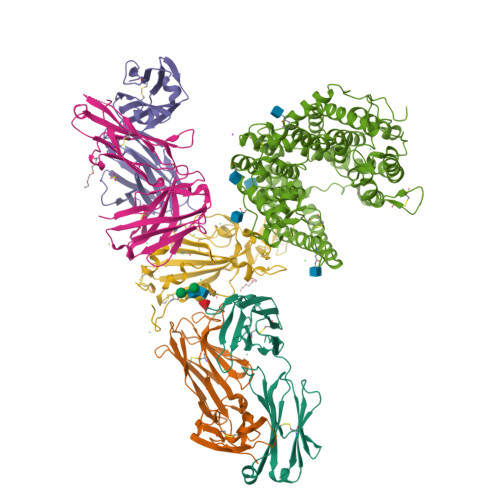
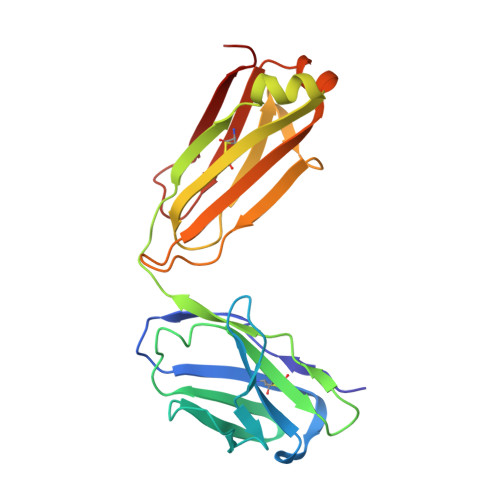
Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Thomson, E.C., Rosen, L.E., Shepherd, J.G., Spreafico, R., da Silva Filipe, A., Wojcechowskyj, J.A., Davis, C., Piccoli, L., Pascall, D.J., Dillen, J., Lytras, S., Czudnochowski, N., Shah, R., Meury, M., Jesudason, N., De Marco, A., Li, K., Bassi, J., O'Toole, A., Pinto, D., Colquhoun, R.M., Culap, K., Jackson, B., Zatta, F., Rambaut, A., Jaconi, S., Sreenu, V.B., Nix, J., Zhang, I., Jarrett, R.F., Glass, W.G., Beltramello, M., Nomikou, K., Pizzuto, M., Tong, L., Cameroni, E., Croll, T.I., Johnson, N., Di Iulio, J., Wickenhagen, A., Ceschi, A., Harbison, A.M., Mair, D., Ferrari, P., Smollett, K., Sallusto, F., Carmichael, S., Garzoni, C., Nichols, J., Galli, M., Hughes, J., Riva, A., Ho, A., Schiuma, M., Semple, M.G., Openshaw, P.J.M., Fadda, E., Baillie, J.K., Chodera, J.D., Rihn, S.J., Lycett, S.J., Virgin, H.W., Telenti, A., Corti, D., Robertson, D.L., Snell, G.(2021) Cell 184: 1171-1187.e20
- PubMed: 33621484
- DOI: https://doi.org/10.1016/j.cell.2021.01.037
- Primary Citation of Related Structures:
7L0N - PubMed Abstract:
SARS-CoV-2 can mutate and evade immunity, with consequences for efficacy of emerging vaccines and antibody therapeutics. Here, we demonstrate that the immunodominant SARS-CoV-2 spike (S) receptor binding motif (RBM) is a highly variable region of S and provide epidemiological, clinical, and molecular characterization of a prevalent, sentinel RBM mutation, N439K. We demonstrate N439K S protein has enhanced binding affinity to the hACE2 receptor, and N439K viruses have similar in vitro replication fitness and cause infections with similar clinical outcomes as compared to wild type. We show the N439K mutation confers resistance against several neutralizing monoclonal antibodies, including one authorized for emergency use by the US Food and Drug Administration (FDA), and reduces the activity of some polyclonal sera from persons recovered from infection. Immune evasion mutations that maintain virulence and fitness such as N439K can emerge within SARS-CoV-2 S, highlighting the need for ongoing molecular surveillance to guide development and usage of vaccines and therapeutics.
Organizational Affiliation:
MRC-University of Glasgow Centre for Virus Research, University of Glasgow, Glasgow G61 1QH, UK; Department of Clinical Research, London School of Hygiene and Tropical Medicine, London WC1E 7HT, UK.