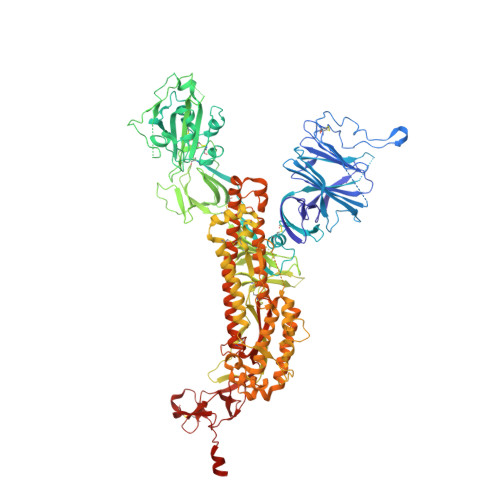
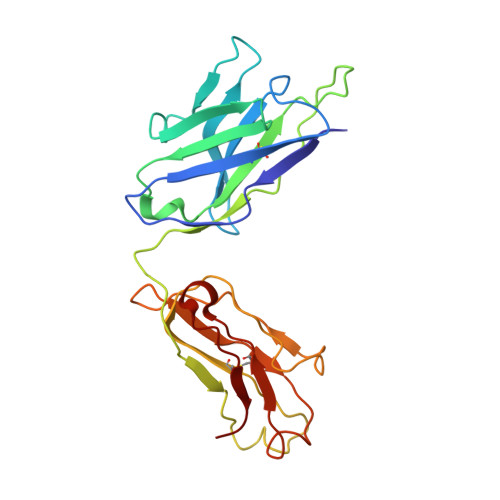
In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies.
Li, D., Edwards, R.J., Manne, K., Martinez, D.R., Schafer, A., Alam, S.M., Wiehe, K., Lu, X., Parks, R., Sutherland, L.L., Oguin 3rd, T.H., McDanal, C., Perez, L.G., Mansouri, K., Gobeil, S.M.C., Janowska, K., Stalls, V., Kopp, M., Cai, F., Lee, E., Foulger, A., Hernandez, G.E., Sanzone, A., Tilahun, K., Jiang, C., Tse, L.V., Bock, K.W., Minai, M., Nagata, B.M., Cronin, K., Gee-Lai, V., Deyton, M., Barr, M., Von Holle, T., Macintyre, A.N., Stover, E., Feldman, J., Hauser, B.M., Caradonna, T.M., Scobey, T.D., Rountree, W., Wang, Y., Moody, M.A., Cain, D.W., DeMarco, C.T., Denny, T.N., Woods, C.W., Petzold, E.W., Schmidt, A.G., Teng, I.T., Zhou, T., Kwong, P.D., Mascola, J.R., Graham, B.S., Moore, I.N., Seder, R., Andersen, H., Lewis, M.G., Montefiori, D.C., Sempowski, G.D., Baric, R.S., Acharya, P., Haynes, B.F., Saunders, K.O.(2021) Cell 184: 4203
- PubMed: 34242577
- DOI: https://doi.org/10.1016/j.cell.2021.06.021
- Primary Citation of Related Structures:
7LAA, 7LAB, 7LCN, 7LD1 - PubMed Abstract:
SARS-CoV-2-neutralizing antibodies (NAbs) protect against COVID-19. A concern regarding SARS-CoV-2 antibodies is whether they mediate disease enhancement. Here, we isolated NAbs against the receptor-binding domain (RBD) or the N-terminal domain (NTD) of SARS-CoV-2 spike from individuals with acute or convalescent SARS-CoV-2 or a history of SARS-CoV infection. Cryo-electron microscopy of RBD and NTD antibodies demonstrated function-specific modes of binding. Select RBD NAbs also demonstrated Fc receptor-γ (FcγR)-mediated enhancement of virus infection in vitro, while five non-neutralizing NTD antibodies mediated FcγR-independent in vitro infection enhancement. However, both types of infection-enhancing antibodies protected from SARS-CoV-2 replication in monkeys and mice. Three of 46 monkeys infused with enhancing antibodies had higher lung inflammation scores compared to controls. One monkey had alveolar edema and elevated bronchoalveolar lavage inflammatory cytokines. Thus, while in vitro antibody-enhanced infection does not necessarily herald enhanced infection in vivo, increased lung inflammation can rarely occur in SARS-CoV-2 antibody-infused macaques.
Organizational Affiliation:
Duke Human Vaccine Institute, Duke University School of Medicine, Durham, NC 27710, USA; Department of Medicine, Duke University School of Medicine, Durham, NC 27710, USA.