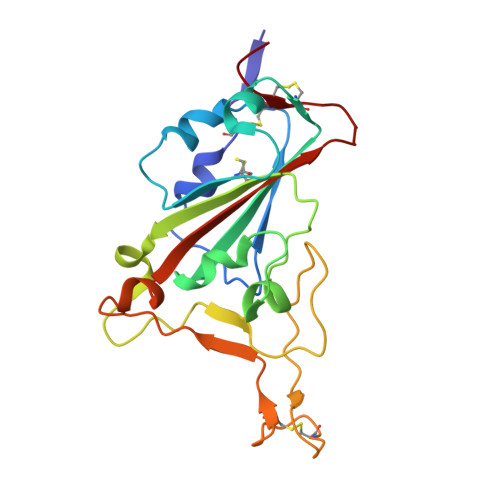
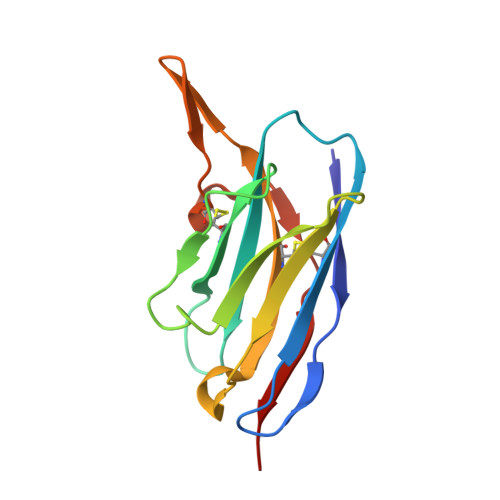
SARS-CoV-2 receptor binding domain in complex with WNb-2
Pymm, P., Adair, A., Chan, L.J., Cooney, J.P., Mordant, F.L., Allison, C.C., Lopez, E., Haycroft, E., O'Neill, M.T., Tan, L.L., Dietrich, M.H., Drew, D., Doerflinger, M., Dengler, M., Scott, N.E., Wheatley, A.K., Gherardin, N.A., Venugopal, H., Cromer, D., Davenport, M.P., Pickering, R., Godfrey, D.I., Purcell, D.J., Kent, S.J., Chung, A.W., Subbarao, K., Pellegrini, M., Glukhova, A., Tham, W.H.(2021) Proc Natl Acad Sci U S A